Os03g0729400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, unclassifiable transcript.


FiT-DB / Search/ Help/ Sample detail
|
Os03g0729400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, unclassifiable transcript.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 323.59 | 0.845 | 3.03 | -0.187 | 0.178 | 0.784 | 0.0149 | -2.83 | -0.153 | 9.6 | 90.66 | 1475 | -- | -- |
| 335.56 | 0.853 | 3 | -0.518 | 0.153 | 0.774 | 0.0386 | -- | -0.202 | 9.9 | 91.94 | 1457 | -- | -- |
| 323.07 | 0.837 | 3.03 | -0.218 | 0.169 | 0.831 | -- | -2.76 | -0.164 | 9.78 | 91.32 | 1466 | -- | -- |
| 335.52 | 0.853 | 3 | -0.499 | 0.16 | 0.764 | -- | -- | -0.204 | 9.31 | 91.84 | 1450 | -- | -- |
| 324.76 | 0.839 | 3.03 | -0.225 | -- | 0.824 | -- | -2.71 | -0.165 | -- | 91.44 | 1466 | -- | -- |
| 345.21 | 0.871 | 3.03 | -0.372 | 0.16 | -- | 0.111 | -- | -0.333 | 8.98 | -- | -- | -- | -- |
| 345.25 | 0.87 | 3.03 | -0.368 | 0.159 | -- | -- | -- | -0.332 | 9.13 | -- | -- | -- | -- |
| 336.97 | 0.855 | 3 | -0.501 | -- | 0.764 | -- | -- | -0.204 | -- | 91.84 | 1447 | -- | -- |
| 341.77 | 0.861 | 2.99 | -- | 0.159 | 0.641 | -- | -- | -0.173 | 9.21 | 92 | 1425 | -- | -- |
| 346.72 | 0.87 | 3.03 | -0.368 | -- | -- | -- | -- | -0.332 | -- | -- | -- | -- | -- |
| 348.82 | 0.874 | 3.02 | -- | 0.159 | -- | -- | -- | -0.291 | 9.31 | -- | -- | -- | -- |
| 343 | 0.863 | 3 | -- | -- | 0.662 | -- | -- | -0.178 | -- | 91.59 | 1245 | -- | -- |
| 350.3 | 0.874 | 3.02 | -- | -- | -- | -- | -- | -0.291 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram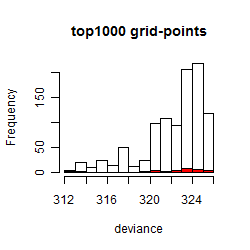 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 312.94 | wind | 9 | 30 | > th | dose dependent | rect. | 11 | 1 |
| 4 | 313.55 | wind | 9 | 30 | > th | dose independent | rect. | 11 | 1 |
| 137 | 319.79 | radiation | 30 | 30 | > th | dose dependent | rect. | 15 | 1 |
| 155 | 320.03 | radiation | 30 | 30 | > th | dose independent | rect. | 15 | 1 |
| 171 | 320.08 | wind | 9 | 30 | > th | dose dependent | sin | 6 | NA |
| 223 | 320.79 | humidity | 60 | 90 | < th | dose independent | rect. | 14 | 3 |
| 278 | 321.09 | humidity | 60 | 90 | < th | dose independent | rect. | 14 | 1 |
| 279 | 321.19 | humidity | 60 | 10 | < th | dose independent | rect. | 15 | 1 |
| 379 | 322.09 | wind | 9 | 30 | < th | dose independent | sin | 5 | NA |
| 395 | 322.37 | wind | 1 | 90 | < th | dose dependent | rect. | 15 | 1 |
| 435 | 322.73 | temperature | 30 | 10 | > th | dose independent | sin | 11 | NA |
| 437 | 322.93 | wind | 9 | 30 | > th | dose independent | sin | 5 | NA |
| 457 | 323.01 | humidity | 90 | 1440 | > th | dose independent | rect. | 1 | 22 |
| 545 | 323.45 | humidity | 60 | 10 | < th | dose dependent | rect. | 15 | 1 |
| 551 | 323.50 | humidity | 90 | 1440 | > th | dose independent | rect. | 9 | 23 |
| 558 | 323.57 | humidity | 90 | 1440 | > th | dose independent | rect. | 15 | 23 |
| 581 | 323.64 | humidity | 90 | 1440 | > th | dose independent | rect. | 7 | 23 |
| 592 | 323.72 | humidity | 90 | 1440 | > th | dose independent | no | NA | NA |
| 621 | 323.84 | humidity | 90 | 1440 | < th | dose independent | rect. | 1 | 20 |
| 651 | 323.94 | humidity | 90 | 1440 | > th | dose independent | rect. | 20 | 23 |
| 699 | 324.16 | humidity | 90 | 1440 | < th | dose independent | rect. | 15 | 22 |
| 714 | 324.26 | humidity | 90 | 1440 | < th | dose independent | rect. | 20 | 23 |
| 747 | 324.38 | humidity | 90 | 1440 | < th | dose independent | rect. | 9 | 23 |
| 770 | 324.44 | humidity | 90 | 1440 | < th | dose independent | no | NA | NA |
| 825 | 324.71 | humidity | 90 | 1440 | < th | dose independent | rect. | 7 | 23 |
| 851 | 324.85 | wind | 1 | 90 | < th | dose independent | rect. | 15 | 1 |
| 899 | 325.09 | humidity | 60 | 10 | < th | dose dependent | sin | 14 | NA |
| 934 | 325.27 | humidity | 90 | 1440 | > th | dose dependent | rect. | 4 | 19 |
| 967 | 325.42 | temperature | 30 | 90 | > th | dose dependent | sin | 11 | NA |
| 988 | 325.54 | humidity | 90 | 1440 | > th | dose dependent | sin | 21 | NA |