Os03g0679300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Shaggy-related protein kinase eta (EC 2.7.1.-) (ASK-eta) (BRASSINOSTEROID-INSENSITIVE 2) (ULTRACURVATA1).
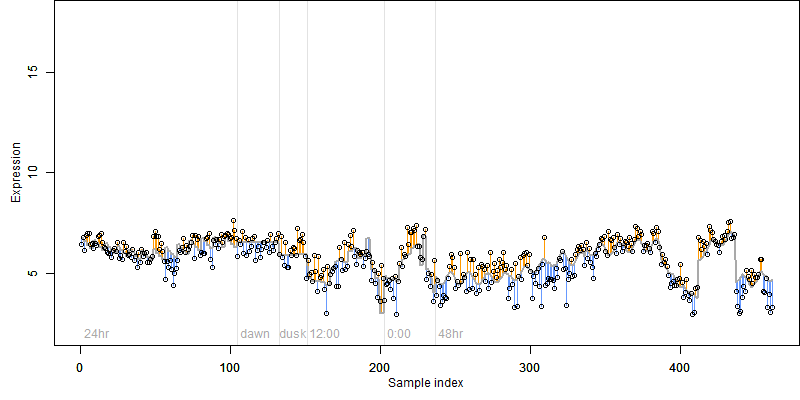
FiT-DB / Search/ Help/ Sample detail
|
Os03g0679300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Shaggy-related protein kinase eta (EC 2.7.1.-) (ASK-eta) (BRASSINOSTEROID-INSENSITIVE 2) (ULTRACURVATA1).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
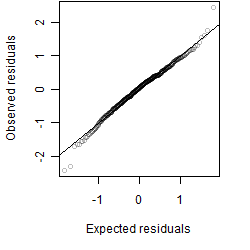
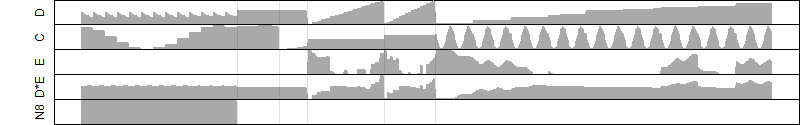
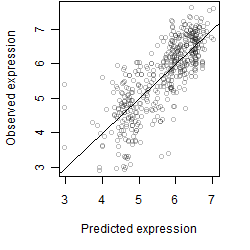 __
__
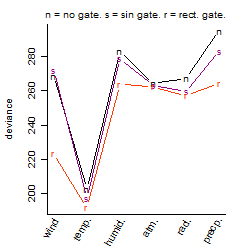
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 186.89 | 0.642 | 5.43 | 1.63 | 0.64 | -2.23 | -0.537 | -5 | 0.504 | 5.43 | 22.1 | 2080 | -- | -- |
| 243.04 | 0.726 | 5.64 | 0.0477 | 0.617 | -1.89 | -0.449 | -- | 0.0257 | 5.44 | 23.7 | 2222 | -- | -- |
| 186.78 | 0.637 | 5.43 | 1.58 | 0.641 | -2.29 | -- | -5.08 | 0.503 | 5.31 | 21.91 | 2058 | -- | -- |
| 243.51 | 0.727 | 5.65 | 0.0553 | 0.575 | -1.99 | -- | -- | -0.0258 | 6.33 | 23.63 | 2230 | -- | -- |
| 208.31 | 0.672 | 5.44 | 1.6 | -- | -2.29 | -- | -5.06 | 0.521 | -- | 22 | 2101 | -- | -- |
| 398.52 | 0.936 | 5.46 | 0.905 | 0.613 | -- | -0.874 | -- | 0.824 | 4.91 | -- | -- | -- | -- |
| 400.14 | 0.937 | 5.46 | 0.906 | 0.608 | -- | -- | -- | 0.828 | 4.44 | -- | -- | -- | -- |
| 253.91 | 0.742 | 5.66 | 0.0201 | -- | -2.09 | -- | -- | -0.0333 | -- | 23.7 | 2591 | -- | -- |
| 243.59 | 0.727 | 5.65 | -- | 0.582 | -2 | -- | -- | -0.0345 | 6.32 | 23.62 | 2230 | -- | -- |
| 419.92 | 0.958 | 5.46 | 0.93 | -- | -- | -- | -- | 0.844 | -- | -- | -- | -- | -- |
| 421.78 | 0.961 | 5.48 | -- | 0.625 | -- | -- | -- | 0.727 | 4.41 | -- | -- | -- | -- |
| 253.71 | 0.742 | 5.65 | -- | -- | -2.09 | -- | -- | 0.00726 | -- | 23.88 | 2598 | -- | -- |
| 442.73 | 0.982 | 5.49 | -- | -- | -- | -- | -- | 0.741 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram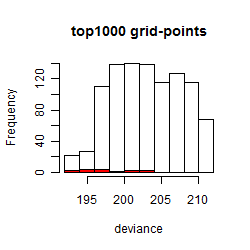 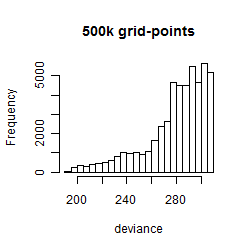
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 192.10 | temperature | 20 | 1440 | > th | dose dependent | rect. | 22 | 9 |
| 11 | 193.14 | temperature | 20 | 1440 | > th | dose dependent | rect. | 23 | 2 |
| 23 | 194.01 | temperature | 25 | 720 | < th | dose dependent | rect. | 4 | 20 |
| 38 | 195.40 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 7 |
| 41 | 195.51 | temperature | 10 | 1440 | > th | dose dependent | rect. | 22 | 4 |
| 48 | 195.83 | temperature | 10 | 1440 | > th | dose dependent | rect. | 23 | 2 |
| 79 | 196.66 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 9 |
| 112 | 197.24 | temperature | 25 | 1440 | < th | dose dependent | sin | 12 | NA |
| 133 | 197.54 | temperature | 25 | 1440 | < th | dose dependent | rect. | 19 | 17 |
| 237 | 199.17 | temperature | 25 | 720 | < th | dose dependent | rect. | 9 | 20 |
| 308 | 200.16 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 1 |
| 314 | 200.34 | temperature | 20 | 1440 | > th | dose dependent | sin | 10 | NA |
| 441 | 202.05 | temperature | 10 | 1440 | > th | dose dependent | sin | 10 | NA |
| 478 | 202.56 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |