Os03g0646300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Cyclic nucleotide-gated channel A (Fragment).

FiT-DB / Search/ Help/ Sample detail
|
Os03g0646300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Cyclic nucleotide-gated channel A (Fragment).
|
|
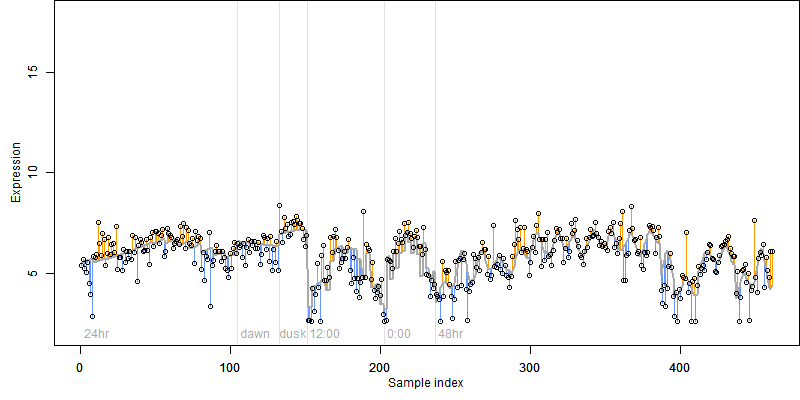
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
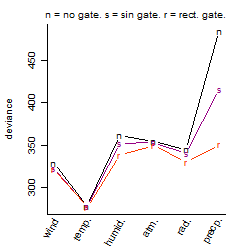
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 278.23 | 0.784 | 5.98 | -1.15 | 1.18 | -4.4 | -0.543 | 0.454 | -0.567 | 20.5 | 25.79 | 1433 | -- | -- |
| 278.4 | 0.777 | 5.97 | -1.06 | 1.19 | -4.44 | -0.545 | -- | -0.542 | 20.5 | 25.57 | 1433 | -- | -- |
| 278.85 | 0.778 | 6 | -1.22 | 1.17 | -4.28 | -- | 0.949 | -0.599 | 20.5 | 26.85 | 1428 | -- | -- |
| 279.32 | 0.778 | 5.97 | -1.04 | 1.18 | -4.44 | -- | -- | -0.539 | 20.6 | 25.57 | 1433 | -- | -- |
| 280.31 | 0.78 | 5.97 | -1.23 | -- | -4.49 | -- | 1.47 | -0.589 | -- | 28.81 | 736 | -- | -- |
| 549 | 1.1 | 5.75 | 0.293 | 1.16 | -- | -0.277 | -- | 0.421 | 21.1 | -- | -- | -- | -- |
| 549.26 | 1.1 | 5.75 | 0.302 | 1.16 | -- | -- | -- | 0.422 | 21.1 | -- | -- | -- | -- |
| 284.01 | 0.785 | 5.93 | -0.962 | -- | -4.5 | -- | -- | -0.531 | -- | 28.48 | 736 | -- | -- |
| 303.51 | 0.811 | 5.93 | -- | 1.17 | -3.88 | -- | -- | -0.342 | 20.4 | 26.02 | 1381 | -- | -- |
| 626.47 | 1.17 | 5.72 | 0.305 | -- | -- | -- | -- | 0.415 | -- | -- | -- | -- | -- |
| 551.67 | 1.1 | 5.76 | -- | 1.16 | -- | -- | -- | 0.388 | 21.1 | -- | -- | -- | -- |
| 305.54 | 0.814 | 5.89 | -- | -- | -4.14 | -- | -- | -0.348 | -- | 28.77 | 724 | -- | -- |
| 628.93 | 1.17 | 5.73 | -- | -- | -- | -- | -- | 0.381 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance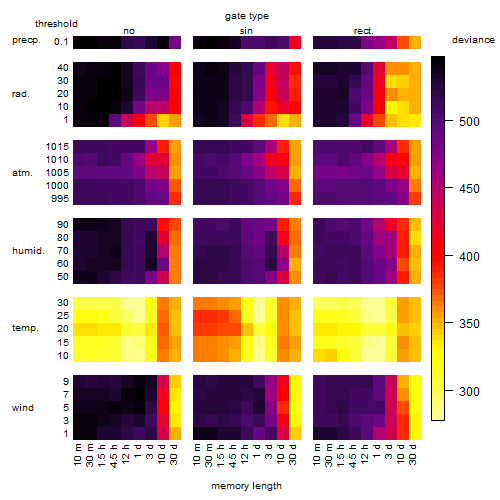
|
Histogram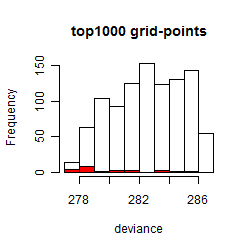 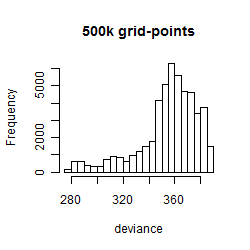
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 277.30 | temperature | 30 | 720 | < th | dose dependent | rect. | 15 | 23 |
| 2 | 277.43 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 21 |
| 3 | 277.47 | temperature | 25 | 1440 | < th | dose dependent | rect. | 17 | 9 |
| 6 | 277.74 | temperature | 25 | 1440 | < th | dose dependent | rect. | 15 | 21 |
| 15 | 278.07 | temperature | 25 | 1440 | < th | dose dependent | sin | 15 | NA |
| 24 | 278.31 | temperature | 25 | 1440 | < th | dose dependent | rect. | 6 | 23 |
| 25 | 278.32 | temperature | 25 | 1440 | < th | dose dependent | rect. | 10 | 23 |
| 37 | 278.57 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 23 |
| 45 | 278.64 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 23 |
| 47 | 278.66 | temperature | 25 | 1440 | < th | dose dependent | no | NA | NA |
| 48 | 278.68 | temperature | 30 | 1440 | < th | dose dependent | rect. | 15 | 18 |
| 69 | 278.91 | temperature | 25 | 1440 | < th | dose dependent | rect. | 10 | 19 |
| 88 | 279.09 | temperature | 30 | 720 | < th | dose dependent | rect. | 24 | 23 |
| 186 | 280.12 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 16 |
| 207 | 280.33 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 2 |
| 266 | 280.98 | temperature | 10 | 1440 | > th | dose dependent | sin | 12 | NA |
| 290 | 281.20 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 10 |
| 319 | 281.43 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 23 |
| 331 | 281.53 | temperature | 10 | 720 | > th | dose dependent | rect. | 20 | 23 |
| 577 | 283.24 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 6 |
| 582 | 283.26 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 2 |
| 597 | 283.35 | temperature | 10 | 720 | > th | dose dependent | rect. | 3 | 23 |
| 737 | 284.49 | temperature | 10 | 720 | > th | dose dependent | no | NA | NA |
| 914 | 285.78 | temperature | 10 | 1440 | > th | dose dependent | rect. | 7 | 23 |