Os03g0632800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Zinc finger, RING-type domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os03g0632800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Zinc finger, RING-type domain containing protein.
|
|
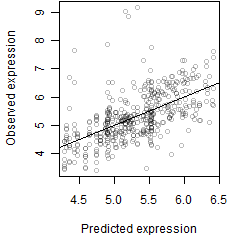
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
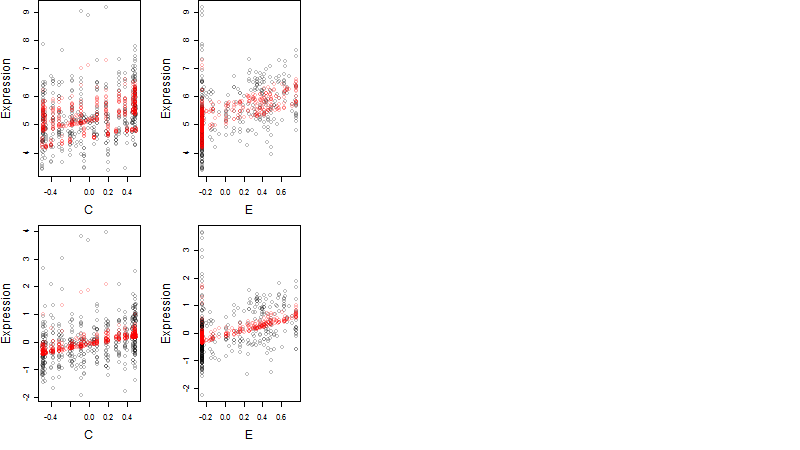
Dependence on each variable
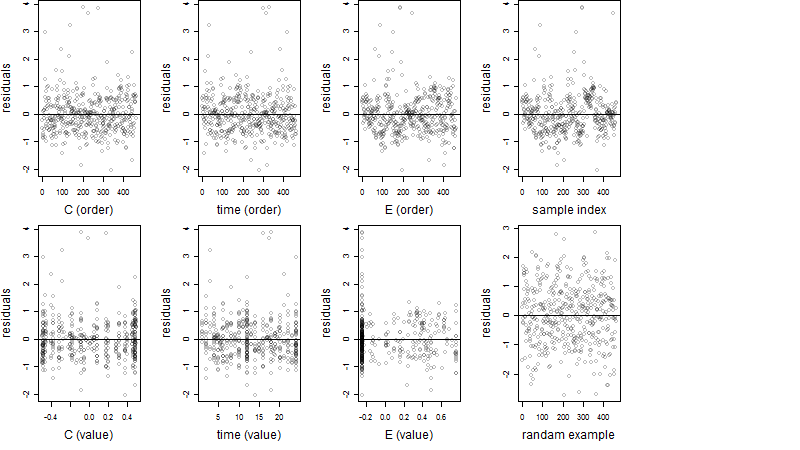
Residual plot
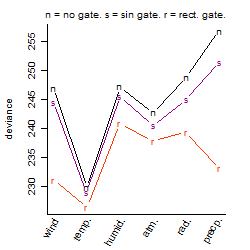
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 225.21 | 0.705 | 5.44 | 0.255 | 0.639 | 1.05 | 0.197 | 1.1 | -0.582 | 11.5 | 22.36 | 1778 | -- | -- |
| 228.07 | 0.703 | 5.4 | 0.647 | 0.635 | 1.07 | 0.195 | -- | -0.488 | 11.5 | 22.21 | 1777 | -- | -- |
| 225.21 | 0.699 | 5.44 | 0.298 | 0.639 | 1.11 | -- | 1.16 | -0.574 | 11.5 | 22.2 | 1785 | -- | -- |
| 228.24 | 0.704 | 5.4 | 0.66 | 0.637 | 1.07 | -- | -- | -0.488 | 11.5 | 22.21 | 1768 | -- | -- |
| 247.69 | 0.733 | 5.45 | 0.321 | -- | 1.16 | -- | 1.11 | -0.548 | -- | 22.2 | 2193 | -- | -- |
| 271.04 | 0.772 | 5.48 | 0.236 | 0.653 | -- | 0.14 | -- | -0.857 | 11.4 | -- | -- | -- | -- |
| 271.12 | 0.771 | 5.48 | 0.242 | 0.652 | -- | -- | -- | -0.857 | 11.4 | -- | -- | -- | -- |
| 250.52 | 0.737 | 5.41 | 0.672 | -- | 1.12 | -- | -- | -0.454 | -- | 22.41 | 2200 | -- | -- |
| 238.09 | 0.719 | 5.43 | -- | 0.637 | 0.89 | -- | -- | -0.606 | 11.4 | 22.4 | 1742 | -- | -- |
| 297.79 | 0.806 | 5.5 | 0.224 | -- | -- | -- | -- | -0.863 | -- | -- | -- | -- | -- |
| 272.66 | 0.772 | 5.49 | -- | 0.65 | -- | -- | -- | -0.883 | 11.4 | -- | -- | -- | -- |
| 261.03 | 0.752 | 5.44 | -- | -- | 0.942 | -- | -- | -0.578 | -- | 22.6 | 2119 | -- | -- |
| 299.12 | 0.807 | 5.51 | -- | -- | -- | -- | -- | -0.888 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance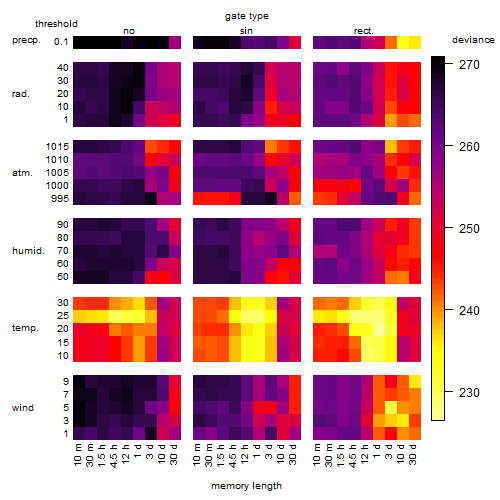
|
Histogram 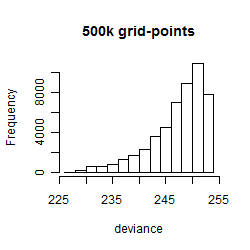
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 226.50 | temperature | 20 | 1440 | > th | dose dependent | rect. | 3 | 1 |
| 14 | 228.13 | temperature | 25 | 720 | < th | dose dependent | rect. | 10 | 20 |
| 24 | 228.43 | temperature | 25 | 270 | < th | dose dependent | rect. | 24 | 23 |
| 29 | 228.55 | temperature | 25 | 720 | < th | dose dependent | rect. | 13 | 17 |
| 73 | 229.03 | temperature | 25 | 270 | < th | dose dependent | rect. | 21 | 23 |
| 74 | 229.05 | temperature | 25 | 720 | < th | dose dependent | sin | 14 | NA |
| 96 | 229.23 | temperature | 10 | 720 | > th | dose dependent | rect. | 17 | 14 |
| 145 | 229.75 | temperature | 25 | 270 | < th | dose dependent | rect. | 15 | 23 |
| 153 | 229.81 | temperature | 25 | 1440 | < th | dose dependent | rect. | 3 | 2 |
| 165 | 229.87 | temperature | 25 | 270 | < th | dose dependent | no | NA | NA |
| 239 | 230.30 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 22 |
| 258 | 230.41 | temperature | 10 | 1440 | > th | dose dependent | rect. | 21 | 9 |
| 363 | 230.76 | temperature | 20 | 4320 | > th | dose independent | rect. | 22 | 1 |
| 419 | 230.93 | temperature | 20 | 1440 | < th | dose independent | rect. | 5 | 1 |
| 490 | 231.17 | wind | 5 | 4320 | > th | dose independent | rect. | 20 | 6 |
| 506 | 231.22 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 2 |
| 553 | 231.43 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 23 |
| 587 | 231.53 | wind | 5 | 4320 | < th | dose independent | rect. | 20 | 6 |
| 667 | 231.77 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 897 | 232.61 | temperature | 10 | 1440 | > th | dose dependent | rect. | 22 | 1 |
| 997 | 233.02 | temperature | 10 | 1440 | > th | dose dependent | sin | 11 | NA |