Os03g0281300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : phosphotransferase system, PEP-utilising enzyme, N-terminal domain containing protein.

FiT-DB / Search/ Help/ Sample detail
|
Os03g0281300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : phosphotransferase system, PEP-utilising enzyme, N-terminal domain containing protein.
|
|
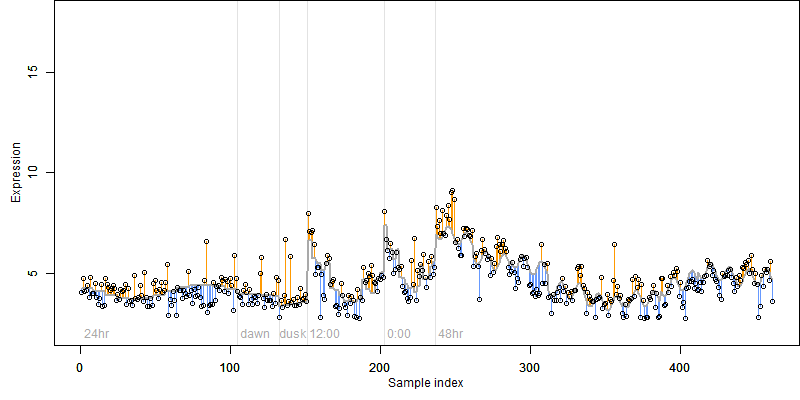
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

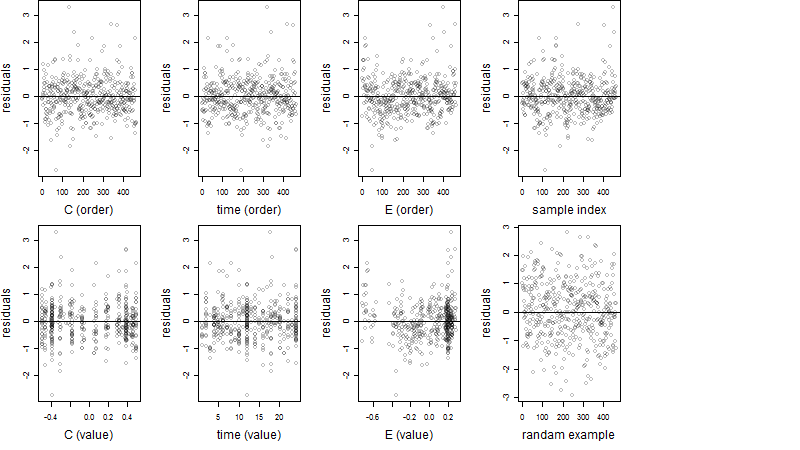
Dependence on each variable

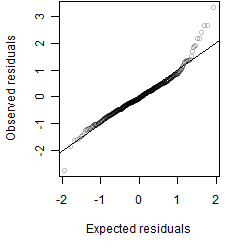
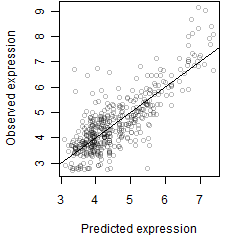
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 196.36 | 0.658 | 4.4 | 0.464 | 0.775 | -2.99 | 0.201 | 4.92 | 0.115 | 2.17 | 14.7 | 5254 | -- | -- |
| 211.1 | 0.677 | 4.52 | -0.234 | 0.739 | -3.43 | 0.523 | -- | 0.182 | 2.08 | 12.3 | 10434 | -- | -- |
| 196.5 | 0.653 | 4.4 | 0.473 | 0.774 | -2.98 | -- | 4.97 | 0.119 | 2.22 | 14.77 | 5254 | -- | -- |
| 210.68 | 0.676 | 4.5 | -0.179 | 0.738 | -3.54 | -- | -- | 0.237 | 2.65 | 8.13 | 10356 | -- | -- |
| 222.27 | 0.694 | 4.29 | 0.81 | -- | -2.95 | -- | 4.89 | 0.494 | -- | -7.971 | 10415 | -- | -- |
| 417.28 | 0.958 | 4.75 | -2.29 | 0.633 | -- | 0.194 | -- | -0.889 | 2.66 | -- | -- | -- | -- |
| 417.4 | 0.957 | 4.75 | -2.29 | 0.635 | -- | -- | -- | -0.89 | 2.73 | -- | -- | -- | -- |
| 241.12 | 0.723 | 4.47 | 0.0361 | -- | -3.49 | -- | -- | 0.342 | -- | 3.435 | 11856 | -- | -- |
| 211.28 | 0.677 | 4.48 | -- | 0.752 | -3.65 | -- | -- | 0.276 | 2.6 | 7.873 | 10385 | -- | -- |
| 440.82 | 0.981 | 4.75 | -2.27 | -- | -- | -- | -- | -0.874 | -- | -- | -- | -- | -- |
| 556.11 | 1.1 | 4.69 | -- | 0.59 | -- | -- | -- | -0.634 | 2.69 | -- | -- | -- | -- |
| 241.14 | 0.723 | 4.47 | -- | -- | -3.47 | -- | -- | 0.332 | -- | 7.699 | 11855 | -- | -- |
| 576.38 | 1.12 | 4.69 | -- | -- | -- | -- | -- | -0.622 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 191.77 | temperature | 10 | 14400 | > th | dose dependent | rect. | 15 | 15 |
| 4 | 191.81 | temperature | 10 | 14400 | > th | dose dependent | rect. | 19 | 7 |
| 5 | 191.81 | temperature | 10 | 14400 | > th | dose dependent | rect. | 18 | 9 |
| 7 | 191.82 | temperature | 30 | 14400 | < th | dose dependent | rect. | 19 | 7 |
| 14 | 191.85 | temperature | 10 | 14400 | > th | dose dependent | sin | 13 | NA |
| 23 | 191.89 | temperature | 10 | 14400 | > th | dose dependent | rect. | 21 | 1 |
| 31 | 191.93 | temperature | 30 | 14400 | < th | dose dependent | rect. | 21 | 1 |
| 65 | 192.10 | temperature | 30 | 14400 | < th | dose dependent | sin | 13 | NA |
| 251 | 192.90 | temperature | 10 | 14400 | > th | dose dependent | rect. | 19 | 22 |
| 285 | 193.00 | temperature | 10 | 14400 | > th | dose dependent | no | NA | NA |
| 310 | 193.08 | temperature | 10 | 14400 | > th | dose dependent | rect. | 24 | 23 |
| 320 | 193.10 | temperature | 10 | 14400 | > th | dose dependent | rect. | 5 | 23 |
| 356 | 193.22 | temperature | 10 | 4320 | > th | dose dependent | rect. | 11 | 2 |
| 485 | 193.64 | temperature | 10 | 4320 | > th | dose dependent | rect. | 10 | 4 |
| 509 | 193.69 | temperature | 30 | 14400 | < th | dose dependent | no | NA | NA |
| 514 | 193.70 | temperature | 15 | 4320 | > th | dose dependent | rect. | 7 | 8 |
| 536 | 193.74 | temperature | 30 | 14400 | < th | dose dependent | rect. | 21 | 23 |
| 538 | 193.75 | temperature | 30 | 4320 | < th | dose dependent | rect. | 7 | 7 |
| 567 | 193.80 | temperature | 30 | 14400 | < th | dose dependent | rect. | 24 | 23 |
| 574 | 193.82 | temperature | 30 | 14400 | < th | dose dependent | rect. | 5 | 23 |
| 609 | 193.90 | temperature | 30 | 14400 | < th | dose dependent | rect. | 5 | 2 |
| 629 | 193.94 | temperature | 10 | 14400 | > th | dose dependent | rect. | 5 | 2 |
| 707 | 194.07 | temperature | 30 | 4320 | < th | dose dependent | rect. | 6 | 11 |