Os03g0280800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : UDP-glucuronic acid decarboxylase (EC 4.1.1.35).

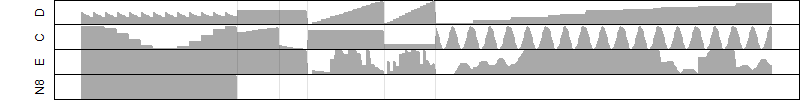
FiT-DB / Search/ Help/ Sample detail
|
Os03g0280800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : UDP-glucuronic acid decarboxylase (EC 4.1.1.35).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
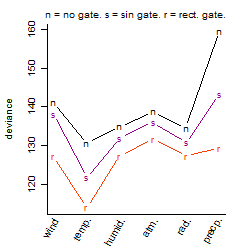
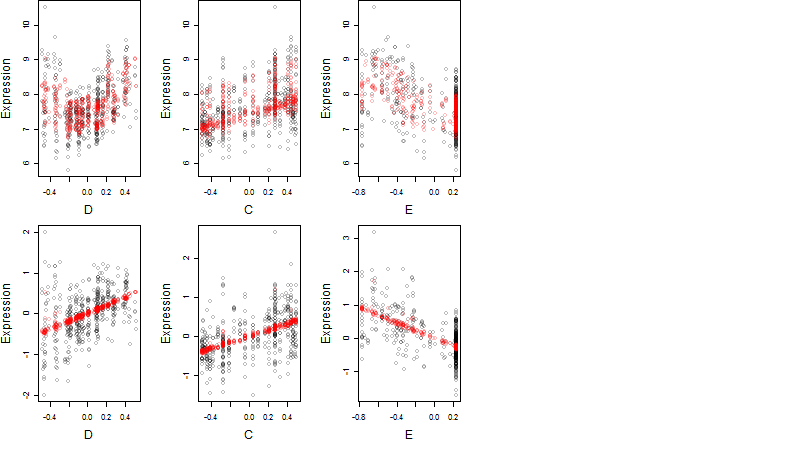
Dependence on each variable
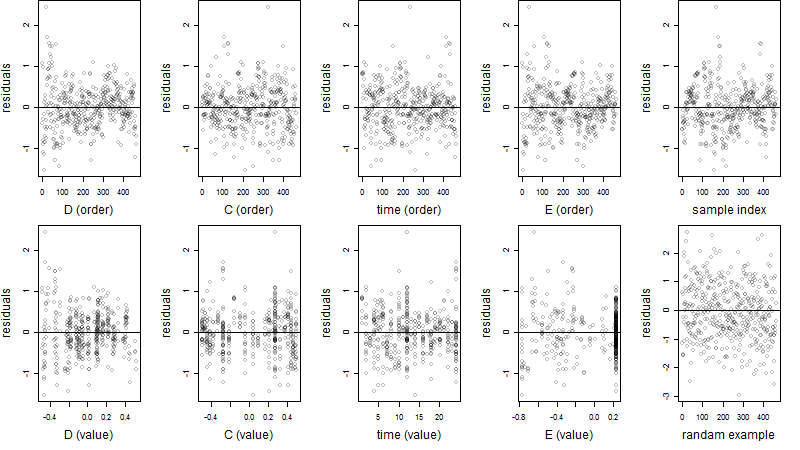
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 122.94 | 0.521 | 7.57 | 1.2 | 0.766 | -1.29 | 0.561 | 0.135 | 0.196 | 8.7 | 21.5 | 2090 | -- | -- |
| 122.96 | 0.516 | 7.58 | 1.15 | 0.77 | -1.28 | 0.574 | -- | 0.191 | 8.64 | 21.6 | 2125 | -- | -- |
| 121.22 | 0.513 | 7.56 | 1.12 | 0.806 | -1.13 | -- | 0.497 | 0.221 | 8.4 | 22.39 | 1245 | -- | -- |
| 121.46 | 0.513 | 7.56 | 0.988 | 0.774 | -1.15 | -- | -- | 0.209 | 8.3 | 22.2 | 1147 | -- | -- |
| 146.09 | 0.563 | 7.59 | 1 | -- | -1.2 | -- | 0.0964 | 0.209 | -- | 22.2 | 776 | -- | -- |
| 172.26 | 0.615 | 7.66 | 0.556 | 0.848 | -- | 0.578 | -- | -0.209 | 8.85 | -- | -- | -- | -- |
| 173.38 | 0.617 | 7.66 | 0.576 | 0.845 | -- | -- | -- | -0.209 | 8.91 | -- | -- | -- | -- |
| 146.12 | 0.563 | 7.59 | 0.969 | -- | -1.2 | -- | -- | 0.202 | -- | 22.21 | 776 | -- | -- |
| 143.5 | 0.558 | 7.62 | -- | 0.735 | -0.914 | -- | -- | 0.00853 | 8.28 | 22.34 | 994 | -- | -- |
| 214.21 | 0.684 | 7.68 | 0.576 | -- | -- | -- | -- | -0.203 | -- | -- | -- | -- | -- |
| 182.13 | 0.631 | 7.68 | -- | 0.846 | -- | -- | -- | -0.273 | 8.85 | -- | -- | -- | -- |
| 169.45 | 0.606 | 7.63 | -- | -- | -1.03 | -- | -- | 0.0433 | -- | 22.27 | 763 | -- | -- |
| 222.96 | 0.697 | 7.7 | -- | -- | -- | -- | -- | -0.267 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 114.12 | temperature | 20 | 1440 | < th | dose independent | rect. | 5 | 1 |
| 2 | 114.60 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 3 | 116.13 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 3 |
| 5 | 117.13 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 3 |
| 17 | 118.66 | temperature | 20 | 720 | > th | dose dependent | rect. | 20 | 13 |
| 39 | 119.97 | temperature | 20 | 4320 | > th | dose independent | rect. | 22 | 1 |
| 45 | 120.22 | temperature | 20 | 1440 | > th | dose dependent | rect. | 1 | 2 |
| 52 | 120.65 | temperature | 25 | 1440 | < th | dose dependent | rect. | 2 | 1 |
| 65 | 121.17 | temperature | 20 | 4320 | < th | dose independent | rect. | 21 | 2 |
| 85 | 121.70 | temperature | 20 | 720 | > th | dose dependent | sin | 9 | NA |
| 90 | 121.76 | temperature | 25 | 720 | < th | dose dependent | rect. | 20 | 11 |
| 95 | 121.84 | temperature | 25 | 720 | < th | dose dependent | rect. | 19 | 13 |
| 131 | 122.23 | temperature | 25 | 720 | < th | dose dependent | sin | 10 | NA |
| 516 | 125.99 | temperature | 20 | 1440 | > th | dose independent | sin | 10 | NA |
| 668 | 126.85 | temperature | 20 | 1440 | < th | dose independent | sin | 10 | NA |
| 745 | 127.26 | radiation | 10 | 4320 | < th | dose independent | rect. | 16 | 14 |
| 747 | 127.26 | wind | 9 | 43200 | > th | dose dependent | rect. | 18 | 13 |
| 762 | 127.32 | humidity | 90 | 43200 | > th | dose independent | rect. | 5 | 1 |
| 829 | 127.70 | humidity | 90 | 43200 | > th | dose dependent | rect. | 5 | 1 |
| 884 | 127.98 | wind | 9 | 43200 | > th | dose dependent | rect. | 18 | 7 |
| 889 | 128.00 | humidity | 90 | 14400 | > th | dose dependent | rect. | 21 | 8 |
| 903 | 128.12 | radiation | 10 | 4320 | < th | dose independent | rect. | 16 | 1 |
| 918 | 128.19 | humidity | 90 | 14400 | > th | dose dependent | rect. | 22 | 6 |
| 952 | 128.33 | radiation | 10 | 4320 | < th | dose independent | rect. | 16 | 7 |
| 959 | 128.35 | radiation | 10 | 4320 | < th | dose independent | rect. | 16 | 5 |
| 963 | 128.36 | radiation | 10 | 4320 | < th | dose independent | rect. | 16 | 3 |
| 971 | 128.39 | humidity | 90 | 14400 | > th | dose dependent | rect. | 24 | 1 |
| 976 | 128.40 | radiation | 10 | 4320 | < th | dose independent | rect. | 16 | 11 |