Os03g0277700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF26 domain containing protein.

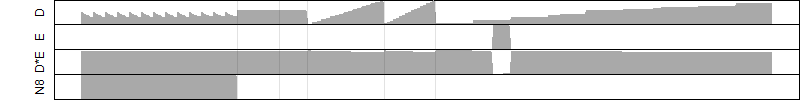
FiT-DB / Search/ Help/ Sample detail
|
Os03g0277700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF26 domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
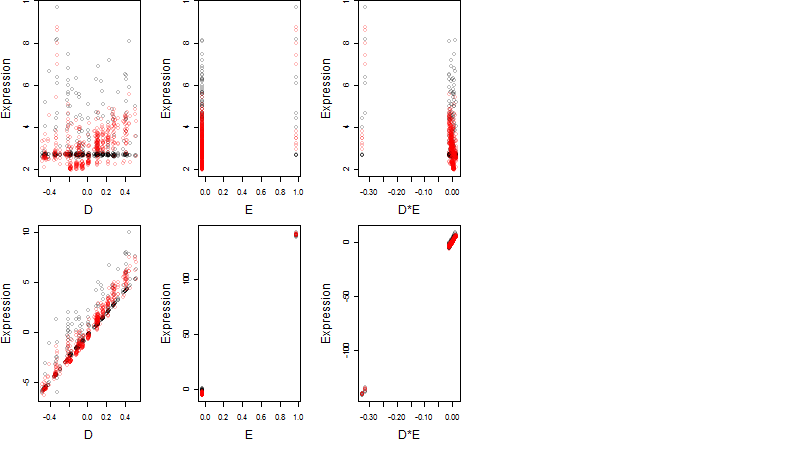
Dependence on each variable
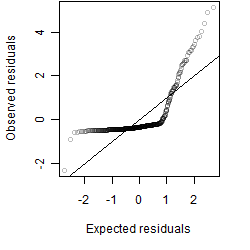
Residual plot

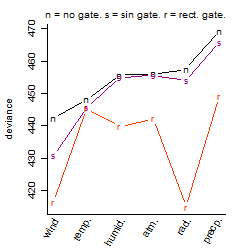
Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = < th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 414.74 | 0.949 | 6.85 | 11.4 | 0.0528 | 144 | -1.05 | 433 | -0.166 | 0.572 | 1 | 1442 | 11 | 1 |
| 484.03 | 1.03 | 3.17 | -0.132 | 0.252 | -0.328 | -1.55 | -- | -0.263 | 3.65 | 1 | 1442 | 11 | 1 |
| 418.47 | 0.953 | 6.84 | 11.4 | 0.151 | 144 | -- | 433 | -0.163 | 5.04 | 0.9949 | 1401 | 11 | 0.959 |
| 463.52 | 1 | 3.13 | -0.222 | 0.394 | 1.3 | -- | -- | -0.264 | 4.52 | 0.2635 | 1446 | 11.3 | 0.933 |
| 419.67 | 0.954 | 6.85 | 11.5 | -- | 144 | -- | 434 | -0.159 | -- | 0.7749 | 1401 | 11 | 1.01 |
| 484.14 | 1.03 | 3.17 | -0.128 | 0.253 | -- | -1.55 | -- | -0.261 | 3.62 | -- | -- | -- | -- |
| 490.57 | 1.04 | 3.16 | -0.102 | 0.285 | -- | -- | -- | -0.253 | 5.36 | -- | -- | -- | -- |
| 457.42 | 0.996 | 3.15 | 0.117 | -- | 1.83 | -- | -- | -0.136 | -- | 4.08 | 1451 | 11.3 | 0.817 |
| 457.66 | 0.996 | 3.16 | -- | 0.354 | 1.8 | -- | -- | -0.218 | 4.62 | 1.338 | 1333 | 11.3 | 0.871 |
| 494.82 | 1.04 | 3.17 | -0.0925 | -- | -- | -- | -- | -0.246 | -- | -- | -- | -- | -- |
| 490.85 | 1.04 | 3.16 | -- | 0.284 | -- | -- | -- | -0.242 | 5.38 | -- | -- | -- | -- |
| 457.69 | 0.996 | 3.14 | -- | -- | 1.81 | -- | -- | -0.153 | -- | 4.09 | 1378 | 11.3 | 0.867 |
| 495.04 | 1.04 | 3.16 | -- | -- | -- | -- | -- | -0.236 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance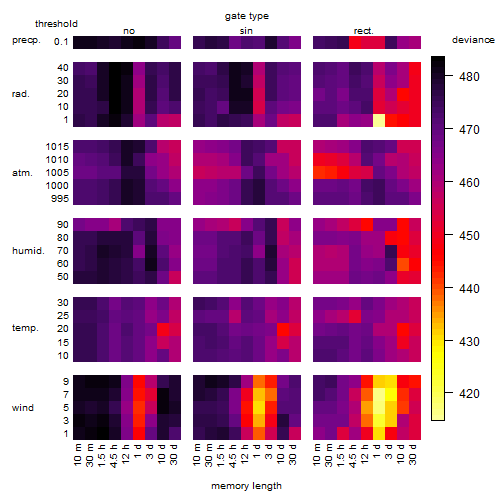
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 414.74 | radiation | 1 | 1440 | < th | dose independent | rect. | 11 | 1 |
| 2 | 414.74 | radiation | 1 | 1440 | < th | dose dependent | rect. | 11 | 1 |
| 3 | 416.27 | wind | 7 | 1440 | > th | dose dependent | rect. | 23 | 2 |
| 6 | 417.84 | wind | 7 | 1440 | > th | dose independent | rect. | 23 | 2 |
| 8 | 418.10 | wind | 5 | 1440 | > th | dose independent | rect. | 21 | 5 |
| 9 | 418.14 | wind | 5 | 1440 | < th | dose independent | rect. | 21 | 5 |
| 10 | 418.36 | wind | 7 | 1440 | < th | dose independent | rect. | 22 | 3 |
| 65 | 423.52 | wind | 3 | 1440 | > th | dose dependent | rect. | 1 | 1 |
| 75 | 423.79 | wind | 7 | 1440 | < th | dose independent | rect. | 21 | 11 |
| 178 | 427.37 | wind | 7 | 4320 | > th | dose independent | rect. | 17 | 3 |
| 209 | 428.36 | wind | 7 | 4320 | > th | dose dependent | rect. | 17 | 3 |
| 210 | 428.37 | wind | 7 | 4320 | < th | dose independent | rect. | 17 | 4 |
| 299 | 430.89 | wind | 5 | 1440 | > th | dose independent | sin | 12 | NA |
| 303 | 430.96 | wind | 5 | 1440 | < th | dose independent | sin | 12 | NA |
| 307 | 431.05 | wind | 9 | 1440 | < th | dose dependent | rect. | 22 | 1 |
| 349 | 431.78 | wind | 5 | 1440 | > th | dose dependent | sin | 12 | NA |
| 408 | 432.56 | wind | 9 | 1440 | < th | dose dependent | rect. | 21 | 6 |
| 427 | 432.97 | wind | 9 | 1440 | < th | dose dependent | rect. | 1 | 2 |
| 449 | 433.28 | wind | 7 | 4320 | > th | dose independent | rect. | 15 | 11 |
| 460 | 433.51 | wind | 7 | 1440 | < th | dose independent | rect. | 14 | 18 |
| 479 | 433.70 | wind | 7 | 1440 | < th | dose independent | rect. | 14 | 16 |
| 493 | 433.85 | wind | 5 | 1440 | > th | dose dependent | rect. | 14 | 18 |
| 501 | 433.91 | wind | 5 | 1440 | > th | dose independent | rect. | 13 | 18 |
| 550 | 434.39 | wind | 7 | 4320 | < th | dose independent | rect. | 15 | 11 |
| 636 | 435.10 | wind | 7 | 4320 | > th | dose dependent | rect. | 15 | 8 |
| 698 | 435.59 | wind | 7 | 4320 | > th | dose dependent | rect. | 15 | 11 |
| 919 | 437.21 | wind | 9 | 1440 | < th | dose dependent | rect. | 13 | 17 |