Os03g0275100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to TA11 protein (Fragment).

FiT-DB / Search/ Help/ Sample detail
|
Os03g0275100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to TA11 protein (Fragment).
|
|
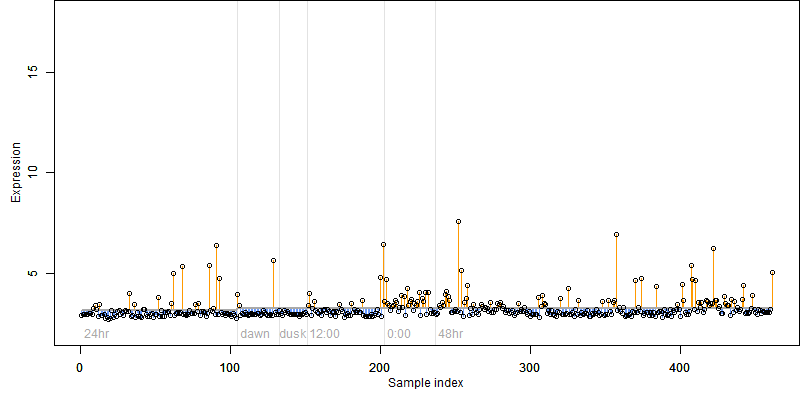
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

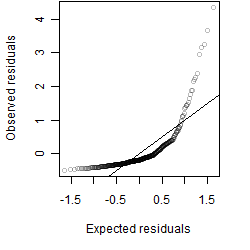
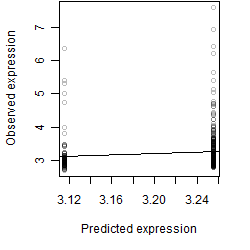
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 136.07 | 0.548 | 3.25 | 0.145 | 0.261 | 0.836 | -0.494 | 0.644 | -0.0332 | 0.463 | 19.3 | 1952 | -- | -- |
| 136.7 | 0.545 | 3.23 | 0.243 | 0.265 | 0.671 | -0.597 | -- | 0.00019 | 0.132 | 19.79 | 1989 | -- | -- |
| 136.8 | 0.545 | 3.25 | 0.134 | 0.283 | 0.864 | -- | 0.77 | -0.0375 | 1.47 | 19.35 | 1952 | -- | -- |
| 137.58 | 0.546 | 3.23 | 0.258 | 0.268 | 0.74 | -- | -- | -0.0111 | 1.48 | 19.36 | 1890 | -- | -- |
| 141.15 | 0.553 | 3.24 | 0.172 | -- | 0.879 | -- | 0.612 | -0.031 | -- | 19.3 | 1787 | -- | -- |
| 145.35 | 0.565 | 3.26 | -0.0674 | 0.228 | -- | -0.601 | -- | -0.15 | 0.176 | -- | -- | -- | -- |
| 146.23 | 0.566 | 3.26 | -0.047 | 0.258 | -- | -- | -- | -0.15 | 1.94 | -- | -- | -- | -- |
| 141.67 | 0.554 | 3.23 | 0.254 | -- | 0.769 | -- | -- | -0.0115 | -- | 19.4 | 1778 | -- | -- |
| 138.89 | 0.549 | 3.24 | -- | 0.276 | 0.64 | -- | -- | -0.0421 | 1.97 | 19.3 | 1856 | -- | -- |
| 150.24 | 0.573 | 3.26 | -0.0363 | -- | -- | -- | -- | -0.144 | -- | -- | -- | -- | -- |
| 146.29 | 0.566 | 3.26 | -- | 0.257 | -- | -- | -- | -0.144 | 1.93 | -- | -- | -- | -- |
| 142.97 | 0.557 | 3.24 | -- | -- | 0.628 | -- | -- | -0.0581 | -- | 19.38 | 1778 | -- | -- |
| 150.28 | 0.572 | 3.26 | -- | -- | -- | -- | -- | -0.14 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 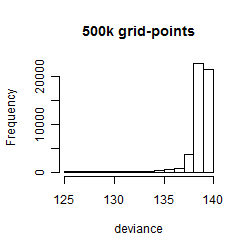
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 125.09 | radiation | 20 | 10 | > th | dose dependent | rect. | 16 | 1 |
| 31 | 125.10 | radiation | 40 | 90 | > th | dose independent | rect. | 15 | 1 |
| 48 | 125.10 | radiation | 40 | 90 | > th | dose dependent | rect. | 15 | 1 |
| 65 | 125.15 | radiation | 20 | 10 | > th | dose independent | rect. | 16 | 1 |
| 255 | 126.82 | wind | 1 | 90 | < th | dose dependent | rect. | 15 | 1 |
| 288 | 127.33 | humidity | 60 | 10 | < th | dose independent | rect. | 15 | 1 |
| 311 | 127.99 | humidity | 60 | 10 | < th | dose dependent | rect. | 14 | 2 |
| 317 | 128.38 | humidity | 60 | 10 | < th | dose independent | rect. | 14 | 5 |
| 352 | 128.50 | humidity | 60 | 10 | < th | dose dependent | rect. | 14 | 5 |
| 384 | 128.81 | humidity | 60 | 90 | < th | dose independent | sin | 14 | NA |
| 385 | 128.81 | wind | 1 | 90 | < th | dose independent | rect. | 15 | 1 |
| 386 | 128.86 | humidity | 60 | 10 | < th | dose independent | sin | 13 | NA |
| 424 | 129.28 | humidity | 60 | 90 | > th | dose independent | sin | 13 | NA |
| 442 | 129.31 | humidity | 60 | 10 | < th | dose dependent | sin | 13 | NA |
| 443 | 129.31 | wind | 3 | 10 | < th | dose dependent | rect. | 16 | 1 |
| 449 | 129.56 | humidity | 60 | 10 | > th | dose independent | sin | 12 | NA |
| 530 | 130.19 | wind | 3 | 10 | < th | dose independent | rect. | 16 | 1 |
| 733 | 132.55 | atmosphere | 1005 | 10 | < th | dose dependent | rect. | 16 | 1 |
| 747 | 132.61 | humidity | 60 | 30 | < th | dose dependent | rect. | 12 | 4 |
| 749 | 132.65 | atmosphere | 1005 | 90 | < th | dose dependent | rect. | 14 | 1 |
| 888 | 134.08 | temperature | 20 | 1440 | < th | dose dependent | rect. | 22 | 1 |
| 890 | 134.11 | temperature | 20 | 1440 | < th | dose dependent | rect. | 16 | 8 |
| 917 | 134.16 | humidity | 60 | 10 | < th | dose independent | rect. | 12 | 5 |
| 937 | 134.17 | wind | 1 | 270 | < th | dose dependent | rect. | 5 | 4 |
| 941 | 134.22 | temperature | 20 | 1440 | < th | dose independent | rect. | 16 | 3 |
| 951 | 134.27 | humidity | 60 | 10 | < th | dose independent | rect. | 12 | 7 |