Os03g0237100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to NADPH-dependent codeinone reductase (EC 1.1.1.247).
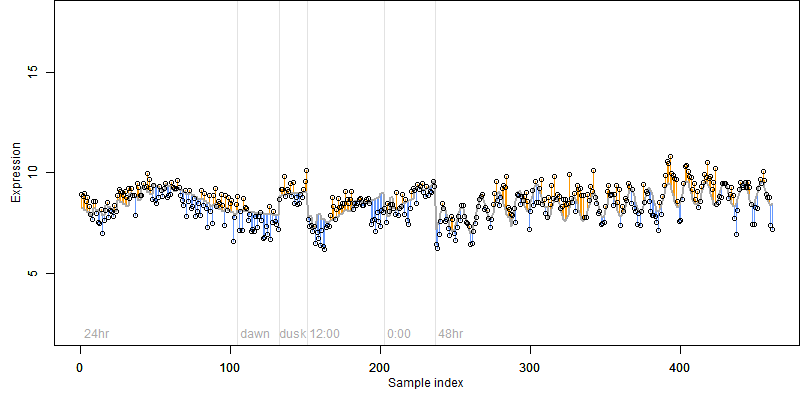

FiT-DB / Search/ Help/ Sample detail
|
Os03g0237100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to NADPH-dependent codeinone reductase (EC 1.1.1.247).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

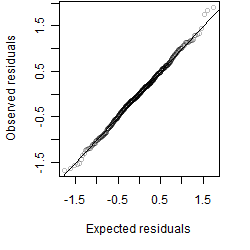
Dependence on each variable

Residual plot
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 160.91 | 0.596 | 8.65 | 2.06 | 1.22 | -1.58 | 0.987 | -3.53 | -0.0291 | 20 | 7.389 | 5965 | -- | -- |
| 171.95 | 0.611 | 8.5 | 1.9 | 1.22 | -1.06 | 0.998 | -- | 0.148 | 20 | 7.399 | 5962 | -- | -- |
| 163.81 | 0.596 | 8.65 | 2.03 | 1.22 | -1.58 | -- | -3.54 | -0.0286 | 20 | 7.337 | 5966 | -- | -- |
| 173.86 | 0.614 | 8.49 | 1.76 | 1.21 | -1 | -- | -- | 0.15 | 19.9 | 7.199 | 5555 | -- | -- |
| 241.75 | 0.724 | 8.6 | 1.85 | -- | -1.6 | -- | -3.5 | -0.00752 | -- | 7.199 | 5229 | -- | -- |
| 190.63 | 0.647 | 8.46 | 1.33 | 1.23 | -- | 0.993 | -- | 0.319 | 20.1 | -- | -- | -- | -- |
| 193.59 | 0.652 | 8.45 | 1.3 | 1.23 | -- | -- | -- | 0.32 | 20.1 | -- | -- | -- | -- |
| 248.52 | 0.734 | 8.44 | 1.75 | -- | -1.19 | -- | -- | 0.217 | -- | 6.7 | 5064 | -- | -- |
| 234.76 | 0.718 | 8.49 | -- | 1.25 | 0.441 | -- | -- | 0.155 | 19.9 | 6.405 | 4010 | -- | -- |
| 276.69 | 0.777 | 8.42 | 1.29 | -- | -- | -- | -- | 0.304 | -- | -- | -- | -- | -- |
| 238.01 | 0.722 | 8.49 | -- | 1.22 | -- | -- | -- | 0.175 | 20.1 | -- | -- | -- | -- |
| 292.78 | 0.797 | 8.5 | -- | -- | 1 | -- | -- | -0.0219 | -- | 4.578 | 47520 | -- | -- |
| 320.37 | 0.835 | 8.46 | -- | -- | -- | -- | -- | 0.161 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 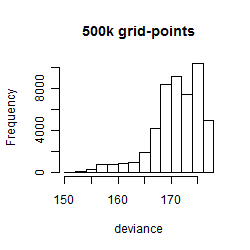
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 151.67 | wind | 7 | 43200 | > th | dose independent | rect. | 17 | 1 |
| 2 | 151.70 | wind | 7 | 43200 | > th | dose dependent | rect. | 15 | 15 |
| 6 | 152.24 | wind | 7 | 43200 | > th | dose dependent | rect. | 17 | 1 |
| 7 | 152.32 | wind | 7 | 43200 | > th | dose independent | rect. | 16 | 14 |
| 8 | 152.34 | wind | 7 | 43200 | < th | dose independent | rect. | 16 | 14 |
| 9 | 152.41 | wind | 7 | 43200 | < th | dose independent | rect. | 17 | 1 |
| 14 | 152.52 | wind | 7 | 43200 | > th | dose independent | rect. | 17 | 3 |
| 29 | 152.94 | wind | 7 | 43200 | > th | dose dependent | rect. | 17 | 3 |
| 33 | 153.04 | wind | 7 | 43200 | > th | dose dependent | rect. | 15 | 8 |
| 52 | 153.38 | wind | 7 | 43200 | > th | dose independent | sin | 14 | NA |
| 55 | 153.41 | wind | 7 | 43200 | < th | dose independent | sin | 14 | NA |
| 60 | 153.50 | wind | 7 | 43200 | < th | dose independent | rect. | 17 | 3 |
| 67 | 153.59 | wind | 7 | 43200 | > th | dose dependent | sin | 14 | NA |
| 71 | 153.72 | wind | 7 | 43200 | > th | dose independent | rect. | 16 | 7 |
| 98 | 153.97 | wind | 9 | 43200 | < th | dose independent | rect. | 15 | 6 |
| 306 | 155.54 | wind | 7 | 43200 | < th | dose independent | rect. | 19 | 1 |
| 501 | 156.51 | wind | 7 | 43200 | > th | dose independent | rect. | 19 | 1 |
| 514 | 156.55 | wind | 7 | 43200 | > th | dose independent | rect. | 2 | 21 |
| 549 | 156.66 | wind | 7 | 43200 | > th | dose independent | no | NA | NA |
| 552 | 156.66 | wind | 7 | 43200 | < th | dose independent | rect. | 2 | 21 |
| 562 | 156.68 | wind | 7 | 43200 | > th | dose independent | rect. | 22 | 23 |
| 596 | 156.74 | wind | 7 | 43200 | > th | dose dependent | rect. | 4 | 18 |
| 601 | 156.76 | wind | 7 | 43200 | < th | dose independent | no | NA | NA |
| 617 | 156.78 | wind | 7 | 43200 | < th | dose independent | rect. | 22 | 23 |
| 666 | 156.88 | wind | 7 | 43200 | > th | dose independent | rect. | 20 | 23 |
| 744 | 157.03 | wind | 7 | 43200 | < th | dose independent | rect. | 20 | 23 |
| 773 | 157.08 | wind | 7 | 43200 | > th | dose dependent | no | NA | NA |
| 802 | 157.13 | wind | 7 | 43200 | > th | dose dependent | rect. | 20 | 23 |
| 924 | 157.52 | wind | 7 | 43200 | < th | dose independent | rect. | 18 | 23 |
| 942 | 157.60 | wind | 7 | 43200 | < th | dose independent | rect. | 16 | 23 |