Os03g0163400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF1677, Oryza sativa family protein.
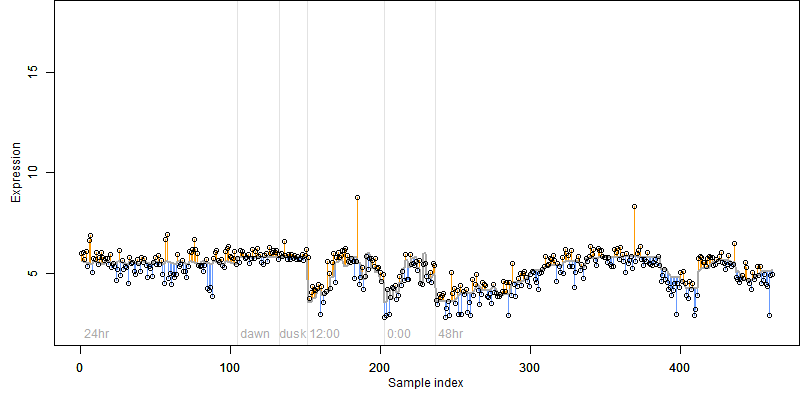

FiT-DB / Search/ Help/ Sample detail
|
Os03g0163400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF1677, Oryza sativa family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

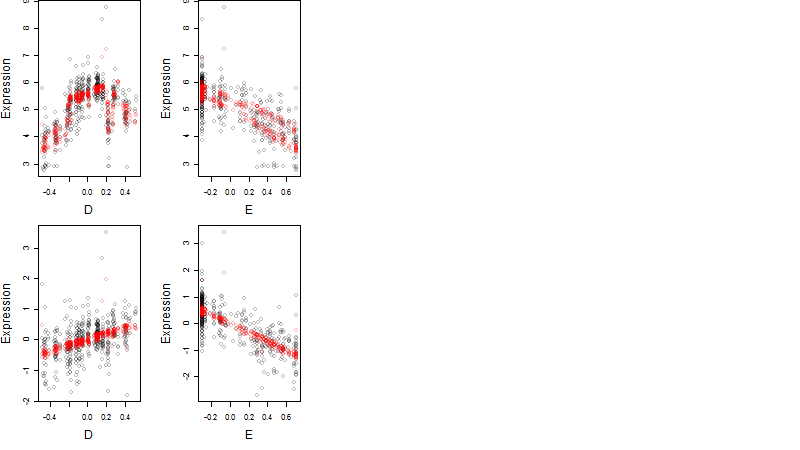
Dependence on each variable
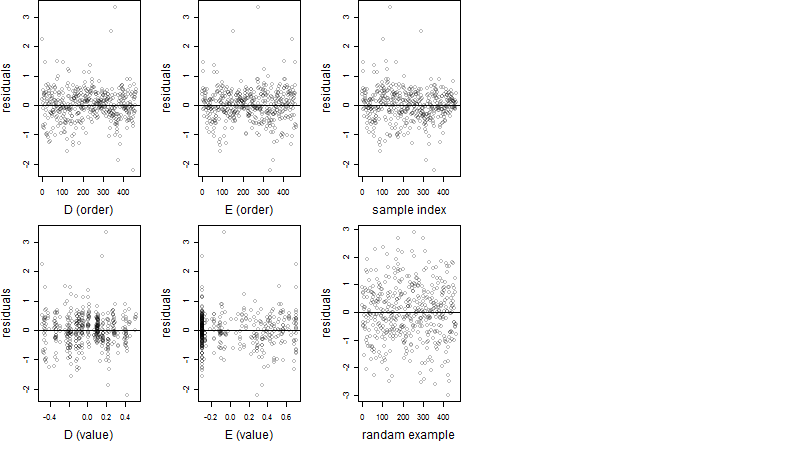
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 126.78 | 0.529 | 5.12 | 1.32 | 0.209 | -1.66 | -0.892 | -0.971 | 0.0213 | 8.79 | 23.1 | 1671 | -- | -- |
| 128.68 | 0.528 | 5.17 | 0.957 | 0.232 | -1.68 | -0.901 | -- | -0.0844 | 8.64 | 23.11 | 1728 | -- | -- |
| 129.22 | 0.529 | 5.12 | 1.28 | 0.233 | -1.66 | -- | -0.977 | 0.0208 | 9.51 | 23.1 | 1755 | -- | -- |
| 131.11 | 0.533 | 5.17 | 0.932 | 0.222 | -1.69 | -- | -- | -0.0877 | 9.43 | 23.3 | 1680 | -- | -- |
| 131.96 | 0.535 | 5.13 | 1.3 | -- | -1.65 | -- | -0.996 | 0.028 | -- | 23.13 | 1642 | -- | -- |
| 249.01 | 0.74 | 5 | 1.63 | 0.204 | -- | -0.818 | -- | 0.623 | 8.61 | -- | -- | -- | -- |
| 251.11 | 0.742 | 5 | 1.61 | 0.206 | -- | -- | -- | 0.624 | 9.32 | -- | -- | -- | -- |
| 133.88 | 0.539 | 5.17 | 0.928 | -- | -1.68 | -- | -- | -0.0885 | -- | 23.28 | 1630 | -- | -- |
| 151.28 | 0.573 | 5.21 | -- | 0.255 | -1.88 | -- | -- | -0.283 | 9.29 | 23.25 | 1782 | -- | -- |
| 253.57 | 0.744 | 5.01 | 1.61 | -- | -- | -- | -- | 0.625 | -- | -- | -- | -- | -- |
| 319.2 | 0.836 | 5.04 | -- | 0.207 | -- | -- | -- | 0.445 | 8.74 | -- | -- | -- | -- |
| 154.36 | 0.579 | 5.21 | -- | -- | -1.89 | -- | -- | -0.273 | -- | 23.22 | 1668 | -- | -- |
| 321.64 | 0.837 | 5.05 | -- | -- | -- | -- | -- | 0.447 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance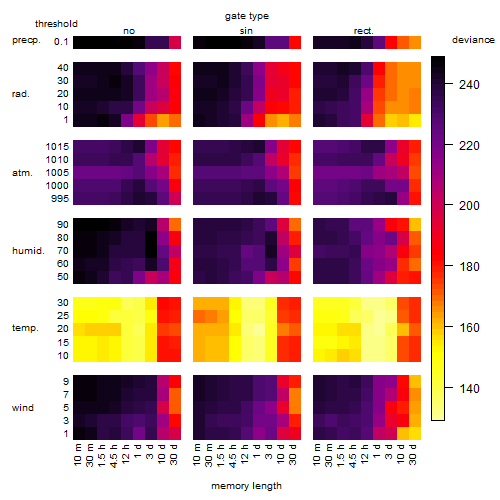
|
Histogram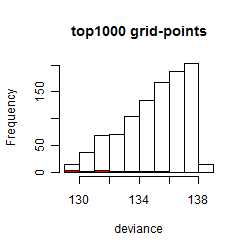 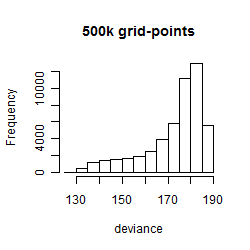
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 129.18 | temperature | 20 | 1440 | > th | dose dependent | rect. | 17 | 14 |
| 11 | 129.86 | temperature | 25 | 1440 | < th | dose dependent | rect. | 18 | 5 |
| 13 | 129.90 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 7 |
| 28 | 130.39 | temperature | 20 | 1440 | > th | dose dependent | sin | 12 | NA |
| 33 | 130.52 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 2 |
| 82 | 131.51 | temperature | 10 | 1440 | > th | dose dependent | rect. | 18 | 5 |
| 90 | 131.62 | temperature | 30 | 720 | < th | dose dependent | rect. | 15 | 18 |
| 96 | 131.70 | temperature | 10 | 1440 | > th | dose dependent | rect. | 19 | 3 |
| 104 | 131.80 | temperature | 10 | 1440 | > th | dose dependent | rect. | 17 | 8 |
| 160 | 132.61 | temperature | 15 | 720 | > th | dose dependent | rect. | 13 | 18 |
| 231 | 133.37 | temperature | 10 | 1440 | > th | dose dependent | rect. | 16 | 13 |
| 238 | 133.42 | temperature | 20 | 1440 | > th | dose dependent | rect. | 4 | 3 |
| 296 | 134.02 | temperature | 30 | 1440 | < th | dose dependent | sin | 14 | NA |
| 458 | 135.23 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 20 |
| 498 | 135.44 | temperature | 30 | 720 | < th | dose dependent | rect. | 9 | 21 |