Os03g0125400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein 147 family protein.
FiT-DB / Search/ Help/ Sample detail
|
Os03g0125400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein 147 family protein.
|
|
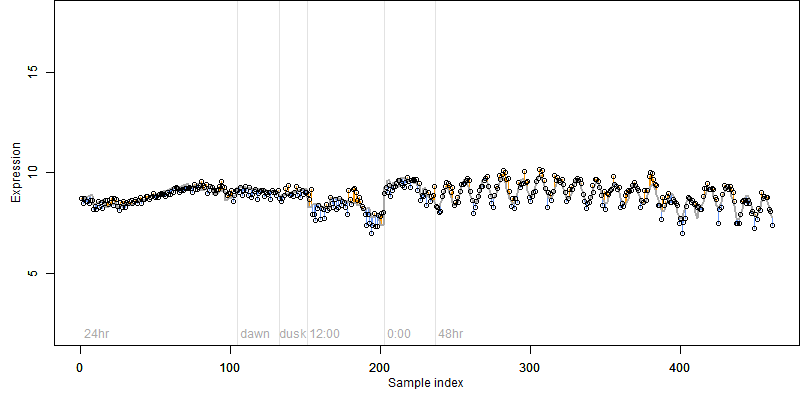
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

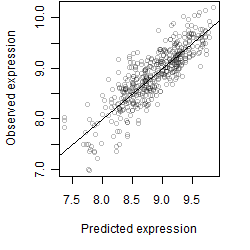 __
__
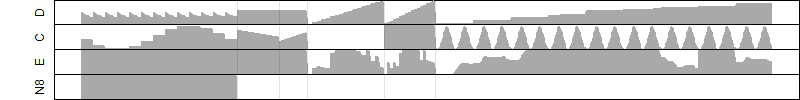
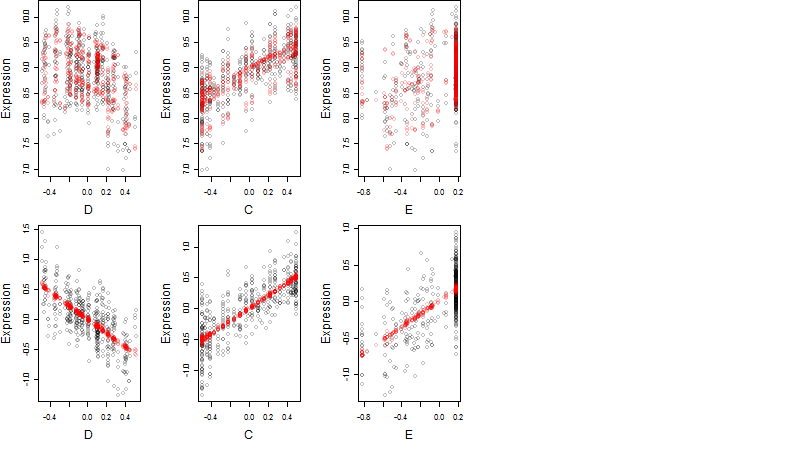
Dependence on each variable
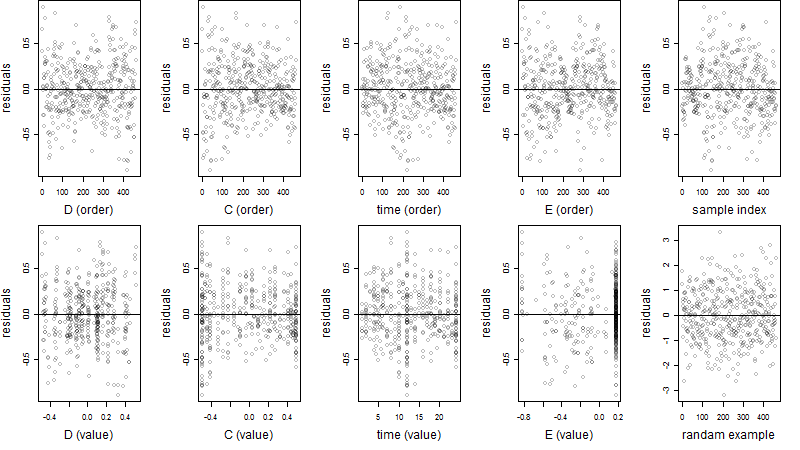
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40.35 | 0.298 | 8.94 | -1.11 | 1.02 | 0.945 | 0.0213 | 0.249 | -0.335 | 0.279 | 21.1 | 1371 | -- | -- |
| 40.32 | 0.296 | 8.95 | -1.14 | 1.01 | 0.89 | 0.0393 | -- | -0.352 | 0.247 | 21.3 | 1323 | -- | -- |
| 40.35 | 0.296 | 8.94 | -1.11 | 1.02 | 0.944 | -- | 0.25 | -0.335 | 0.282 | 21.11 | 1365 | -- | -- |
| 40.32 | 0.296 | 8.95 | -1.14 | 1.01 | 0.889 | -- | -- | -0.35 | 0.273 | 21.3 | 1323 | -- | -- |
| 92.72 | 0.448 | 8.94 | -0.971 | -- | 0.939 | -- | 0.209 | -0.385 | -- | 22.2 | 937 | -- | -- |
| 62.87 | 0.372 | 8.9 | -0.777 | 1.06 | -- | -0.0459 | -- | -0.104 | 0.348 | -- | -- | -- | -- |
| 62.88 | 0.371 | 8.9 | -0.775 | 1.06 | -- | -- | -- | -0.103 | 0.342 | -- | -- | -- | -- |
| 92.71 | 0.448 | 8.95 | -1.06 | -- | 0.959 | -- | -- | -0.412 | -- | 22.3 | 955 | -- | -- |
| 66.37 | 0.379 | 8.9 | -- | 0.996 | 0.595 | -- | -- | -0.141 | 0.221 | 22.3 | 1266 | -- | -- |
| 133.62 | 0.54 | 8.88 | -0.739 | -- | -- | -- | -- | -0.0865 | -- | -- | -- | -- | -- |
| 78.7 | 0.415 | 8.88 | -- | 1.05 | -- | -- | -- | -0.017 | 0.304 | -- | -- | -- | -- |
| 114.51 | 0.498 | 8.91 | -- | -- | 0.842 | -- | -- | -0.239 | -- | 25.1 | 897 | -- | -- |
| 148.03 | 0.568 | 8.86 | -- | -- | -- | -- | -- | -0.00444 | -- | -- | -- | -- | -- |
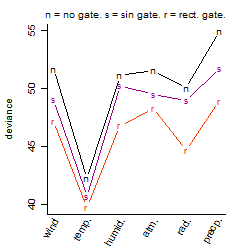
Results of the grid search
Summarized heatmap of deviance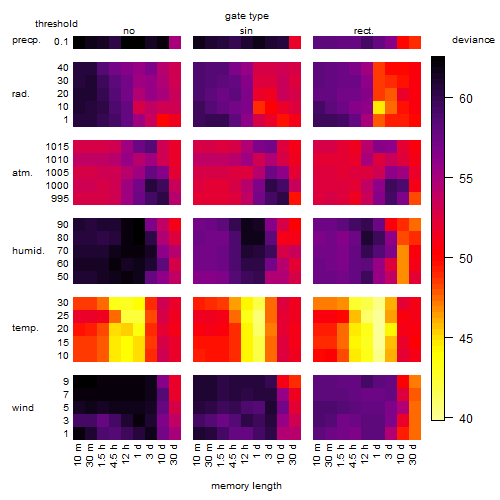
|
Histogram 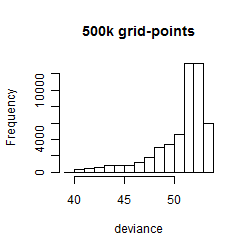
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 39.80 | temperature | 20 | 1440 | > th | dose independent | rect. | 20 | 10 |
| 6 | 40.09 | temperature | 20 | 1440 | < th | dose independent | rect. | 20 | 10 |
| 8 | 40.12 | temperature | 25 | 1440 | < th | dose dependent | rect. | 20 | 8 |
| 12 | 40.15 | temperature | 15 | 1440 | > th | dose dependent | rect. | 23 | 5 |
| 16 | 40.17 | temperature | 30 | 1440 | < th | dose dependent | rect. | 23 | 5 |
| 21 | 40.21 | temperature | 15 | 1440 | > th | dose dependent | rect. | 1 | 2 |
| 42 | 40.31 | temperature | 25 | 1440 | < th | dose dependent | rect. | 21 | 1 |
| 44 | 40.33 | temperature | 30 | 1440 | < th | dose dependent | rect. | 1 | 2 |
| 148 | 40.55 | temperature | 20 | 1440 | < th | dose independent | rect. | 23 | 7 |
| 185 | 40.70 | temperature | 25 | 1440 | < th | dose dependent | sin | 12 | NA |
| 304 | 41.01 | temperature | 20 | 1440 | > th | dose independent | sin | 10 | NA |
| 333 | 41.10 | temperature | 20 | 1440 | > th | dose independent | rect. | 3 | 1 |
| 346 | 41.13 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 20 |
| 361 | 41.21 | temperature | 20 | 1440 | < th | dose independent | rect. | 3 | 1 |
| 421 | 41.34 | temperature | 20 | 1440 | < th | dose independent | sin | 10 | NA |
| 474 | 41.48 | temperature | 20 | 1440 | > th | dose independent | rect. | 23 | 2 |
| 624 | 41.83 | temperature | 15 | 1440 | > th | dose dependent | sin | 11 | NA |
| 627 | 41.83 | temperature | 30 | 720 | < th | dose dependent | rect. | 21 | 20 |
| 697 | 41.98 | temperature | 20 | 1440 | > th | dose independent | rect. | 5 | 1 |
| 850 | 42.25 | temperature | 25 | 720 | < th | dose dependent | no | NA | NA |