Os03g0116000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : 60 kDa inner membrane insertion protein family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os03g0116000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : 60 kDa inner membrane insertion protein family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

Dependence on each variable
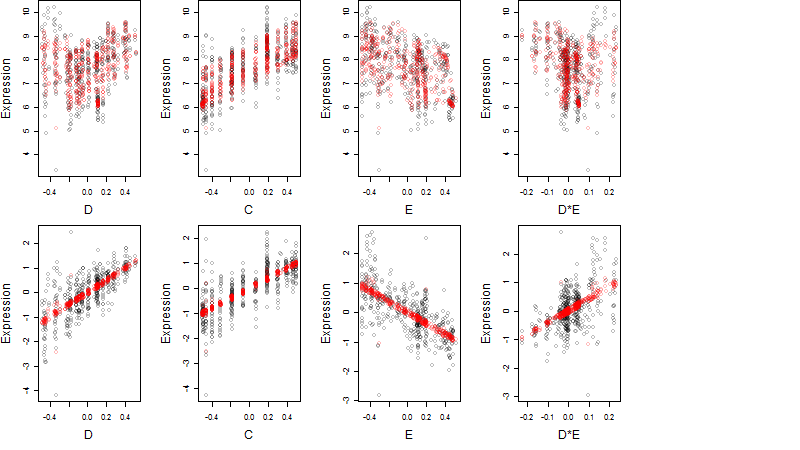
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 173.31 | 0.619 | 7.5 | 2.45 | 2.02 | -1.83 | -0.583 | 4.08 | 0.489 | 16.5 | 21.42 | 1304 | -- | -- |
| 195.46 | 0.651 | 7.62 | 1.56 | 2.1 | -1.65 | -1.12 | -- | 0.22 | 16.1 | 17.77 | 2935 | -- | -- |
| 174.16 | 0.615 | 7.5 | 2.48 | 2.01 | -1.84 | -- | 4.19 | 0.494 | 16.5 | 21.4 | 1295 | -- | -- |
| 194.5 | 0.65 | 7.64 | 1.45 | 2.04 | -1.86 | -- | -- | 0.178 | 16.6 | 15.8 | 2557 | -- | -- |
| 383.4 | 0.912 | 7.47 | 2.56 | -- | -2.24 | -- | 4.53 | 0.504 | -- | 22.16 | 1242 | -- | -- |
| 262.68 | 0.76 | 7.72 | 0.712 | 2.18 | -- | -0.767 | -- | -0.2 | 16.2 | -- | -- | -- | -- |
| 264.26 | 0.761 | 7.72 | 0.704 | 2.17 | -- | -- | -- | -0.204 | 16.3 | -- | -- | -- | -- |
| 398.01 | 0.929 | 7.62 | 1.3 | -- | -1.87 | -- | -- | 0.159 | -- | 21 | 1239 | -- | -- |
| 224.81 | 0.698 | 7.69 | -- | 2.04 | -1.18 | -- | -- | -0.0562 | 16.4 | 21.4 | 1354 | -- | -- |
| 518.89 | 1.06 | 7.71 | 0.617 | -- | -- | -- | -- | -0.262 | -- | -- | -- | -- | -- |
| 277.31 | 0.779 | 7.74 | -- | 2.16 | -- | -- | -- | -0.283 | 16.3 | -- | -- | -- | -- |
| 437.02 | 0.974 | 7.66 | -- | -- | -1.57 | -- | -- | -0.0419 | -- | 21.83 | 1245 | -- | -- |
| 528.95 | 1.07 | 7.72 | -- | -- | -- | -- | -- | -0.33 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 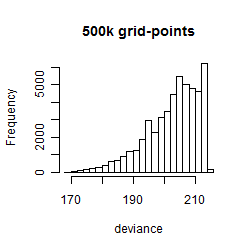
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 169.62 | temperature | 25 | 1440 | > th | dose dependent | rect. | 12 | 7 |
| 6 | 170.30 | temperature | 20 | 720 | > th | dose dependent | rect. | 20 | 18 |
| 10 | 170.58 | temperature | 30 | 720 | < th | dose dependent | sin | 9 | NA |
| 12 | 170.63 | temperature | 30 | 720 | < th | dose dependent | rect. | 19 | 16 |
| 17 | 170.75 | temperature | 20 | 720 | > th | dose dependent | sin | 8 | NA |
| 20 | 170.88 | temperature | 10 | 720 | > th | dose dependent | rect. | 18 | 15 |
| 21 | 170.89 | temperature | 30 | 720 | < th | dose dependent | rect. | 17 | 18 |
| 77 | 172.72 | temperature | 10 | 270 | > th | dose dependent | rect. | 1 | 21 |
| 89 | 173.03 | temperature | 20 | 1440 | > th | dose dependent | rect. | 18 | 3 |
| 98 | 173.14 | temperature | 30 | 720 | < th | dose dependent | rect. | 22 | 19 |
| 154 | 173.79 | temperature | 10 | 720 | > th | dose dependent | sin | 9 | NA |
| 172 | 173.98 | temperature | 10 | 270 | > th | dose dependent | rect. | 2 | 19 |
| 175 | 174.01 | temperature | 20 | 1440 | > th | dose dependent | sin | 16 | NA |
| 252 | 174.98 | temperature | 10 | 720 | > th | dose dependent | rect. | 20 | 17 |
| 439 | 177.11 | temperature | 20 | 1440 | > th | dose dependent | rect. | 2 | 21 |
| 501 | 177.59 | temperature | 20 | 1440 | > th | dose dependent | no | NA | NA |
| 517 | 177.68 | temperature | 25 | 1440 | > th | dose dependent | rect. | 5 | 15 |
| 576 | 177.96 | temperature | 20 | 1440 | > th | dose dependent | rect. | 20 | 23 |
| 708 | 178.85 | humidity | 90 | 43200 | > th | dose dependent | rect. | 22 | 1 |
| 749 | 179.16 | humidity | 90 | 43200 | > th | dose independent | rect. | 22 | 1 |
| 753 | 179.25 | temperature | 10 | 720 | > th | dose dependent | rect. | 12 | 21 |
| 785 | 179.45 | temperature | 25 | 1440 | > th | dose independent | rect. | 14 | 8 |
| 843 | 179.93 | temperature | 25 | 1440 | < th | dose independent | rect. | 15 | 7 |
| 868 | 180.21 | temperature | 25 | 1440 | > th | dose independent | rect. | 9 | 14 |
| 873 | 180.22 | temperature | 10 | 720 | > th | dose dependent | rect. | 10 | 23 |
| 951 | 180.67 | temperature | 20 | 720 | > th | dose dependent | rect. | 1 | 7 |
| 971 | 180.77 | temperature | 25 | 1440 | < th | dose independent | rect. | 9 | 14 |