Os03g0106700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Beta-expansin.

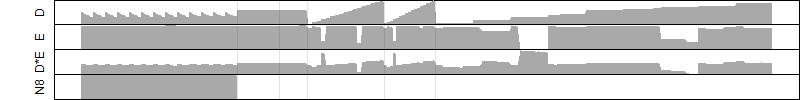
FiT-DB / Search/ Help/ Sample detail
|
Os03g0106700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Beta-expansin.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
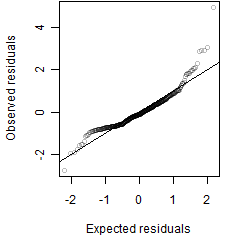
 __
__

Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 256.95 | 0.753 | 3.74 | 0.556 | 0.446 | -1.98 | 1.17 | -9.43 | -0.199 | 8.16 | 6.783 | 4312 | 19 | 2.81 |
| 302.18 | 0.81 | 3.74 | 0.398 | 0.45 | -1.69 | 1.27 | -- | -0.196 | 7.54 | 8.015 | 4318 | 18.9 | 2.55 |
| 260.7 | 0.752 | 3.74 | 0.585 | 0.416 | -1.95 | -- | -9.27 | -0.201 | 8.22 | 6.772 | 4315 | 19 | 2.82 |
| 303.93 | 0.812 | 3.74 | 0.284 | 0.398 | -1.85 | -- | -- | -0.196 | 8.75 | 7.997 | 4318 | 18.9 | 2.6 |
| 269.05 | 0.764 | 3.75 | 0.569 | -- | -1.94 | -- | -9.19 | -0.186 | -- | 6.795 | 4323 | 19 | 2.81 |
| 389.9 | 0.926 | 3.73 | 0.415 | 0.422 | -- | 1.03 | -- | -0.307 | 8.49 | -- | -- | -- | -- |
| 393.31 | 0.929 | 3.73 | 0.447 | 0.42 | -- | -- | -- | -0.307 | 8.51 | -- | -- | -- | -- |
| 296.44 | 0.802 | 3.74 | 0.312 | -- | -2.16 | -- | -- | -0.197 | -- | 9.068 | 4319 | 18.9 | 2.63 |
| 306.64 | 0.816 | 3.74 | -- | 0.415 | -1.93 | -- | -- | -0.195 | 8.33 | 8 | 4318 | 18.9 | 2.51 |
| 403.17 | 0.938 | 3.74 | 0.449 | -- | -- | -- | -- | -0.303 | -- | -- | -- | -- | -- |
| 398.58 | 0.934 | 3.74 | -- | 0.422 | -- | -- | -- | -0.357 | 8.43 | -- | -- | -- | -- |
| 293.37 | 0.798 | 3.73 | -- | -- | -2.07 | -- | -- | -0.208 | -- | 7.722 | 4321 | 18.9 | 1.89 |
| 408.49 | 0.943 | 3.75 | -- | -- | -- | -- | -- | -0.353 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 262.63 | wind | 7 | 4320 | < th | dose independent | rect. | 19 | 3 |
| 2 | 262.95 | wind | 9 | 4320 | < th | dose independent | rect. | 20 | 7 |
| 6 | 263.39 | wind | 5 | 4320 | > th | dose dependent | rect. | 20 | 2 |
| 12 | 264.09 | wind | 7 | 4320 | > th | dose independent | rect. | 19 | 3 |
| 53 | 265.52 | wind | 9 | 4320 | > th | dose independent | rect. | 19 | 7 |
| 136 | 274.18 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 17 | 3 |
| 152 | 276.05 | atmosphere | 995 | 14400 | < th | dose independent | rect. | 16 | 1 |
| 157 | 276.11 | atmosphere | 995 | 14400 | < th | dose dependent | rect. | 16 | 1 |
| 196 | 278.31 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 7 | 13 |
| 350 | 284.01 | radiation | 1 | 14400 | < th | dose dependent | rect. | 22 | 3 |
| 500 | 291.32 | atmosphere | 995 | 14400 | < th | dose independent | sin | 22 | NA |
| 508 | 291.47 | atmosphere | 995 | 14400 | > th | dose independent | sin | 22 | NA |
| 531 | 292.79 | atmosphere | 1005 | 14400 | < th | dose independent | rect. | 5 | 2 |
| 535 | 293.02 | atmosphere | 1005 | 14400 | > th | dose independent | rect. | 6 | 1 |
| 549 | 293.66 | atmosphere | 995 | 14400 | < th | dose dependent | sin | 22 | NA |
| 794 | 297.06 | humidity | 90 | 1440 | < th | dose dependent | rect. | 3 | 1 |
| 939 | 298.02 | atmosphere | 995 | 14400 | > th | dose independent | no | NA | NA |
| 947 | 298.02 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 12 | 23 |
| 948 | 298.02 | atmosphere | 995 | 14400 | > th | dose independent | rect. | 14 | 23 |