Os02g0833000
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Serine/threonine-protein kinase PBS1 (EC 2.7.1.37) (AvrPphB susceptible protein 1).


FiT-DB / Search/ Help/ Sample detail
|
Os02g0833000 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Serine/threonine-protein kinase PBS1 (EC 2.7.1.37) (AvrPphB susceptible protein 1).
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
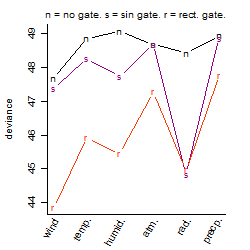
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47.64 | 0.324 | 2.88 | 0.0259 | 0.128 | -0.518 | -0.227 | -0.346 | 0.0218 | 7.02 | 7 | 28 | -- | -- |
| 47.61 | 0.321 | 2.88 | 0.0228 | 0.124 | -0.467 | -0.229 | -- | 0.0234 | 6.95 | 7.2 | 23 | -- | -- |
| 47.73 | 0.322 | 2.88 | 0.0174 | 0.127 | -0.489 | -- | -0.275 | 0.0212 | 7.13 | 6.8 | 31 | -- | -- |
| 47.74 | 0.322 | 2.88 | 0.0159 | 0.122 | -0.449 | -- | -- | 0.0237 | 7.24 | 7.119 | 23 | -- | -- |
| 48.57 | 0.325 | 2.88 | 0.0129 | -- | -0.433 | -- | -0.2 | 0.0235 | -- | 6.878 | 31 | -- | -- |
| 49.33 | 0.329 | 2.88 | -0.0462 | 0.106 | -- | -0.154 | -- | 0.00397 | 8.64 | -- | -- | -- | -- |
| 49.41 | 0.329 | 2.88 | -0.0512 | 0.107 | -- | -- | -- | 0.00392 | 8.59 | -- | -- | -- | -- |
| 48.5 | 0.324 | 2.88 | 0.0176 | -- | -0.414 | -- | -- | 0.0274 | -- | 7.107 | 21 | -- | -- |
| 47.73 | 0.322 | 2.88 | -- | 0.122 | -0.436 | -- | -- | 0.0212 | 7.21 | 7.119 | 21 | -- | -- |
| 50.05 | 0.331 | 2.89 | -0.0509 | -- | -- | -- | -- | 0.00488 | -- | -- | -- | -- | -- |
| 49.48 | 0.329 | 2.88 | -- | 0.107 | -- | -- | -- | 0.00962 | 8.63 | -- | -- | -- | -- |
| 48.51 | 0.324 | 2.88 | -- | -- | -0.405 | -- | -- | 0.0221 | -- | 7.117 | 21 | -- | -- |
| 50.12 | 0.33 | 2.88 | -- | -- | -- | -- | -- | 0.0105 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 43.87 | wind | 7 | 10 | > th | dose dependent | rect. | 12 | 3 |
| 3 | 43.98 | wind | 7 | 90 | > th | dose independent | rect. | 12 | 1 |
| 4 | 44.02 | wind | 7 | 10 | > th | dose independent | rect. | 12 | 3 |
| 55 | 44.85 | radiation | 40 | 30 | > th | dose dependent | sin | 12 | NA |
| 57 | 44.97 | radiation | 40 | 30 | > th | dose dependent | rect. | 13 | 21 |
| 60 | 45.09 | radiation | 40 | 30 | > th | dose independent | sin | 12 | NA |
| 66 | 45.28 | radiation | 40 | 30 | > th | dose independent | rect. | 13 | 21 |
| 71 | 45.45 | radiation | 40 | 10 | > th | dose independent | rect. | 9 | 1 |
| 72 | 45.45 | humidity | 60 | 1440 | > th | dose independent | rect. | 7 | 2 |
| 91 | 45.45 | humidity | 70 | 90 | < th | dose dependent | rect. | 8 | 1 |
| 95 | 45.52 | radiation | 40 | 10 | > th | dose dependent | rect. | 9 | 1 |
| 211 | 45.81 | humidity | 60 | 1440 | < th | dose independent | rect. | 8 | 1 |
| 225 | 45.82 | humidity | 60 | 1440 | > th | dose independent | rect. | 21 | 12 |
| 229 | 45.82 | humidity | 60 | 10 | < th | dose dependent | rect. | 17 | 17 |
| 236 | 45.85 | humidity | 60 | 1440 | < th | dose dependent | rect. | 8 | 1 |
| 309 | 45.92 | temperature | 30 | 90 | > th | dose dependent | rect. | 18 | 16 |
| 313 | 45.93 | humidity | 70 | 90 | < th | dose independent | rect. | 8 | 1 |
| 314 | 45.93 | temperature | 30 | 90 | > th | dose dependent | rect. | 8 | 1 |
| 367 | 45.95 | temperature | 30 | 90 | > th | dose independent | rect. | 8 | 1 |
| 382 | 45.95 | humidity | 60 | 10 | < th | dose independent | rect. | 17 | 18 |
| 401 | 45.96 | humidity | 60 | 90 | < th | dose independent | rect. | 18 | 15 |
| 419 | 45.98 | humidity | 60 | 10 | < th | dose independent | rect. | 9 | 1 |
| 681 | 46.00 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 724 | 46.04 | wind | 1 | 10 | < th | dose independent | rect. | 8 | 2 |
| 730 | 46.07 | temperature | 30 | 90 | > th | dose independent | rect. | 18 | 16 |
| 749 | 46.12 | humidity | 60 | 1440 | > th | dose independent | rect. | 2 | 7 |