Os02g0822700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : AMP-dependent synthetase and ligase domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0822700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : AMP-dependent synthetase and ligase domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
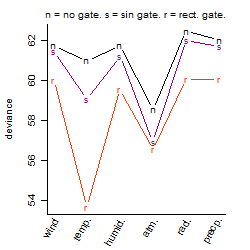
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 57.09 | 0.355 | 2.33 | 3.36 | 0.0301 | -45.5 | 0.222 | 278 | 0.0046 | 14 | 994.9 | 673 | -- | -- |
| 63.14 | 0.373 | 2.88 | 0.0451 | 0.0221 | -0.0395 | -0.19 | -- | -0.00248 | 4.64 | 994.9 | 673 | -- | -- |
| 56.67 | 0.351 | 2.37 | 3.1 | 0.0454 | -45.8 | -- | 280 | 0.0108 | 10.6 | 994.6 | 654 | -- | -- |
| 62.33 | 0.368 | 2.88 | 0.1 | 0.0419 | 0.432 | -- | -- | 0.0299 | 12.3 | 994.5 | 603 | -- | -- |
| 56.76 | 0.351 | 2.38 | 3.08 | -- | -45.9 | -- | 281 | 0.00419 | -- | 994.5 | 658 | -- | -- |
| 63.14 | 0.373 | 2.88 | 0.0449 | 0.023 | -- | -0.188 | -- | -0.00238 | 4.69 | -- | -- | -- | -- |
| 63.17 | 0.372 | 2.88 | 0.0464 | 0.0403 | -- | -- | -- | -0.0013 | 8.26 | -- | -- | -- | -- |
| 62.34 | 0.368 | 2.88 | 0.0741 | -- | 0.496 | -- | -- | 0.0316 | -- | 994.7 | 630 | -- | -- |
| 62.11 | 0.367 | 2.88 | -- | 0.0611 | 0.481 | -- | -- | 0.00511 | 8.55 | 994.6 | 704 | -- | -- |
| 63.26 | 0.372 | 2.88 | 0.0467 | -- | -- | -- | -- | -0.000846 | -- | -- | -- | -- | -- |
| 63.23 | 0.372 | 2.88 | -- | 0.0405 | -- | -- | -- | -0.00647 | 8.17 | -- | -- | -- | -- |
| 62.28 | 0.368 | 2.88 | -- | -- | 0.46 | -- | -- | 0.000421 | -- | 994.6 | 659 | -- | -- |
| 63.32 | 0.371 | 2.88 | -- | -- | -- | -- | -- | -0.00603 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 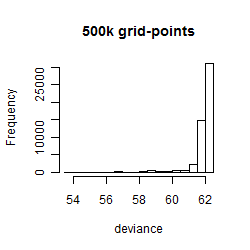
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 53.64 | temperature | 25 | 90 | > th | dose independent | rect. | 1 | 1 |
| 2 | 53.71 | temperature | 25 | 90 | > th | dose dependent | rect. | 1 | 1 |
| 11 | 56.52 | atmosphere | 995 | 720 | < th | dose independent | rect. | 14 | 1 |
| 52 | 56.52 | atmosphere | 995 | 720 | < th | dose dependent | rect. | 14 | 1 |
| 94 | 56.53 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 1 | 1 |
| 102 | 56.53 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 1 | 1 |
| 120 | 56.90 | atmosphere | 995 | 720 | > th | dose independent | sin | 7 | NA |
| 121 | 56.92 | atmosphere | 995 | 720 | < th | dose independent | sin | 6 | NA |
| 129 | 57.24 | atmosphere | 995 | 720 | < th | dose dependent | sin | 7 | NA |
| 168 | 58.02 | atmosphere | 995 | 720 | > th | dose independent | sin | 1 | NA |
| 172 | 58.07 | temperature | 30 | 10 | > th | dose dependent | rect. | 11 | 1 |
| 206 | 58.29 | atmosphere | 1010 | 10 | < th | dose dependent | rect. | 2 | 1 |
| 228 | 58.41 | atmosphere | 995 | 720 | > th | dose independent | sin | 22 | NA |
| 275 | 58.53 | atmosphere | 995 | 720 | > th | dose independent | no | NA | NA |
| 320 | 58.75 | atmosphere | 995 | 720 | < th | dose independent | no | NA | NA |
| 534 | 58.81 | atmosphere | 995 | 720 | > th | dose independent | sin | 16 | NA |
| 544 | 58.90 | atmosphere | 995 | 720 | < th | dose dependent | no | NA | NA |
| 747 | 59.04 | temperature | 30 | 90 | > th | dose independent | rect. | 10 | 2 |
| 770 | 59.07 | temperature | 30 | 10 | > th | dose dependent | sin | 7 | NA |
| 798 | 59.15 | temperature | 30 | 10 | > th | dose independent | rect. | 11 | 1 |
| 826 | 59.26 | temperature | 30 | 30 | < th | dose independent | rect. | 3 | 23 |
| 838 | 59.34 | atmosphere | 995 | 720 | > th | dose independent | sin | 13 | NA |
| 898 | 59.56 | humidity | 90 | 90 | < th | dose independent | rect. | 1 | 2 |