Os02g0786400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Allergen V5/Tpx-1 related family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0786400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Allergen V5/Tpx-1 related family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
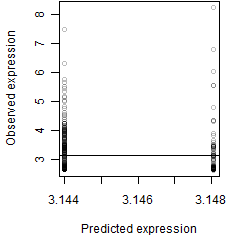
Dependence on each variable

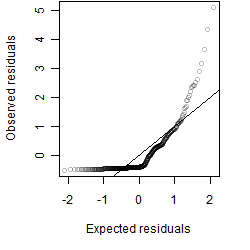
Residual plot

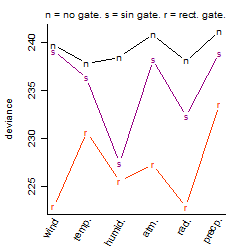
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 237.26 | 0.724 | 3.16 | -0.602 | 0.203 | 0.161 | -0.0773 | 0.999 | -0.0341 | 12.3 | 24.58 | 226 | -- | -- |
| 239.6 | 0.721 | 3.14 | -0.261 | 0.26 | 0.209 | -0.284 | -- | -0.00297 | 12.1 | 28 | 48 | -- | -- |
| 237.18 | 0.717 | 3.16 | -0.625 | 0.2 | 0.159 | -- | 1.04 | -0.0393 | 12.3 | 24.5 | 195 | -- | -- |
| 238.54 | 0.724 | 3.14 | -0.276 | 0.311 | 0.294 | -- | -- | 0.0102 | 12.6 | 27.55 | 63 | -- | -- |
| 239.8 | 0.721 | 3.16 | -0.655 | -- | 0.128 | -- | 1.08 | -0.0551 | -- | 24.51 | 234 | -- | -- |
| 243.43 | 0.731 | 3.15 | -0.35 | 0.179 | -- | -0.182 | -- | -0.0345 | 10.7 | -- | -- | -- | -- |
| 243.51 | 0.731 | 3.15 | -0.359 | 0.185 | -- | -- | -- | -0.0358 | 10 | -- | -- | -- | -- |
| 241.54 | 0.724 | 3.13 | -0.253 | -- | 0.288 | -- | -- | 0.0509 | -- | 22.2 | 852 | -- | -- |
| 240.48 | 0.726 | 3.13 | -- | 0.334 | 0.329 | -- | -- | 0.0452 | 12.8 | 27.52 | 68 | -- | -- |
| 245.57 | 0.732 | 3.15 | -0.361 | -- | -- | -- | -- | -0.036 | -- | -- | -- | -- | -- |
| 246.9 | 0.735 | 3.14 | -- | 0.186 | -- | -- | -- | 0.00411 | 10.2 | -- | -- | -- | -- |
| 243.19 | 0.726 | 3.12 | -- | -- | 0.35 | -- | -- | 0.106 | -- | 22.12 | 851 | -- | -- |
| 249.01 | 0.737 | 3.14 | -- | -- | -- | -- | -- | 0.00405 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance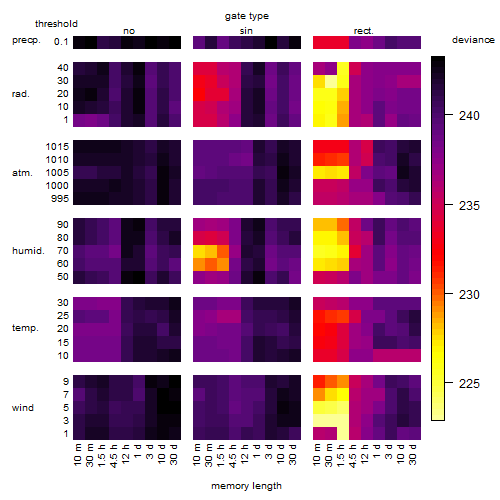
|
Histogram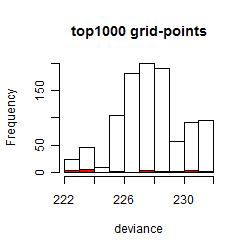 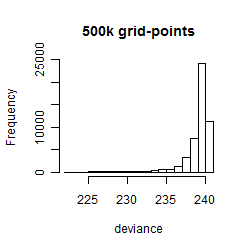
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 222.85 | radiation | 30 | 30 | > th | dose dependent | rect. | 15 | 1 |
| 17 | 222.90 | wind | 1 | 90 | < th | dose dependent | rect. | 15 | 1 |
| 18 | 222.92 | wind | 1 | 90 | < th | dose dependent | rect. | 15 | 3 |
| 27 | 223.06 | wind | 3 | 10 | < th | dose dependent | rect. | 16 | 1 |
| 32 | 223.21 | wind | 3 | 90 | < th | dose independent | rect. | 15 | 1 |
| 35 | 223.36 | wind | 1 | 90 | < th | dose independent | rect. | 14 | 4 |
| 45 | 223.54 | wind | 3 | 10 | < th | dose independent | rect. | 16 | 1 |
| 55 | 223.87 | radiation | 30 | 30 | > th | dose independent | rect. | 15 | 1 |
| 115 | 225.55 | humidity | 70 | 10 | < th | dose dependent | rect. | 15 | 1 |
| 119 | 225.87 | radiation | 1 | 30 | > th | dose dependent | rect. | 15 | 1 |
| 368 | 227.03 | humidity | 60 | 10 | < th | dose independent | rect. | 15 | 1 |
| 376 | 227.25 | atmosphere | 1005 | 90 | < th | dose dependent | rect. | 14 | 1 |
| 378 | 227.28 | atmosphere | 1005 | 10 | < th | dose dependent | rect. | 16 | 1 |
| 388 | 227.43 | humidity | 70 | 10 | < th | dose dependent | sin | 14 | NA |
| 755 | 228.98 | humidity | 60 | 90 | > th | dose independent | sin | 14 | NA |
| 761 | 229.08 | humidity | 60 | 90 | < th | dose independent | sin | 14 | NA |
| 799 | 229.69 | humidity | 60 | 10 | < th | dose independent | sin | 13 | NA |
| 839 | 230.23 | humidity | 60 | 10 | > th | dose independent | sin | 13 | NA |
| 868 | 230.58 | atmosphere | 1005 | 90 | < th | dose independent | rect. | 14 | 1 |
| 872 | 230.65 | temperature | 25 | 90 | > th | dose independent | rect. | 15 | 1 |
| 928 | 231.14 | radiation | 1 | 30 | > th | dose independent | rect. | 15 | 1 |