Os02g0767400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Curculin-like (mannose-binding) lectin domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0767400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Curculin-like (mannose-binding) lectin domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
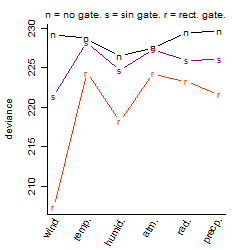
Dependence on each variable

Residual plot

Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = rect.)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 206.23 | 0.675 | 2.94 | 0.0739 | 0.196 | 2.91 | 0.444 | -2.08 | 0.0428 | 3.78 | 1.118 | 7 | 0.997 | 4.07 |
| 206.8 | 0.67 | 2.95 | 0.0785 | 0.193 | 3.22 | 0.301 | -- | 0.0425 | 3.64 | 1.2 | 6 | 1.02 | 4 |
| 206.67 | 0.67 | 2.94 | 0.0717 | 0.199 | 2.95 | -- | -1.41 | 0.0437 | 3.17 | 1.122 | 6 | 1 | 4.02 |
| 206.93 | 0.67 | 2.95 | 0.0651 | 0.204 | 3.18 | -- | -- | 0.0412 | 3.18 | 1.2 | 6 | 1.01 | 4.16 |
| 208.64 | 0.673 | 2.94 | 0.0766 | -- | 3.2 | -- | -1.14 | 0.0527 | -- | 1.092 | 6 | 0.983 | 4.02 |
| 234.73 | 0.718 | 2.95 | -0.0112 | 0.414 | -- | -0.0637 | -- | 0.011 | 3.21 | -- | -- | -- | -- |
| 234.74 | 0.717 | 2.95 | -0.00994 | 0.414 | -- | -- | -- | 0.0114 | 3.22 | -- | -- | -- | -- |
| 208.83 | 0.673 | 2.95 | 0.0705 | -- | 3.3 | -- | -- | 0.0422 | -- | 1.034 | 6 | 0.999 | 4.52 |
| 207.03 | 0.67 | 2.95 | -- | 0.207 | 3.16 | -- | -- | 0.0341 | 3.18 | 1.2 | 6 | 1.01 | 4.6 |
| 244.46 | 0.731 | 2.95 | 0.00765 | -- | -- | -- | -- | 0.0222 | -- | -- | -- | -- | -- |
| 234.74 | 0.717 | 2.95 | -- | 0.414 | -- | -- | -- | 0.0125 | 3.22 | -- | -- | -- | -- |
| 208.96 | 0.673 | 2.95 | -- | -- | 3.27 | -- | -- | 0.035 | -- | 1.018 | 6 | 1 | 4.71 |
| 244.46 | 0.73 | 2.95 | -- | -- | -- | -- | -- | 0.0214 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 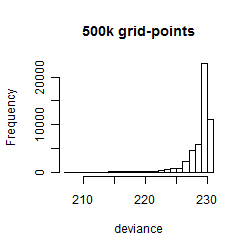
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 207.37 | wind | 1 | 10 | > th | dose dependent | rect. | 1 | 4 |
| 7 | 211.61 | wind | 7 | 720 | > th | dose independent | rect. | 24 | 1 |
| 9 | 211.61 | wind | 5 | 720 | > th | dose dependent | rect. | 1 | 1 |
| 26 | 212.67 | wind | 5 | 10 | > th | dose dependent | rect. | 24 | 9 |
| 29 | 212.82 | wind | 7 | 90 | > th | dose independent | rect. | 24 | 8 |
| 32 | 212.96 | wind | 3 | 30 | > th | dose independent | rect. | 1 | 3 |
| 52 | 213.79 | wind | 5 | 10 | > th | dose independent | rect. | 24 | 8 |
| 90 | 214.40 | wind | 7 | 90 | > th | dose independent | rect. | 24 | 4 |
| 131 | 215.05 | wind | 7 | 10 | > th | dose independent | rect. | 20 | 8 |
| 253 | 215.94 | wind | 7 | 10 | > th | dose dependent | rect. | 20 | 8 |
| 300 | 216.55 | wind | 7 | 720 | > th | dose independent | rect. | 19 | 9 |
| 314 | 216.68 | wind | 7 | 720 | > th | dose independent | rect. | 19 | 13 |
| 357 | 216.95 | wind | 5 | 720 | < th | dose independent | rect. | 20 | 7 |
| 466 | 218.20 | humidity | 60 | 90 | < th | dose dependent | rect. | 9 | 1 |
| 497 | 218.35 | wind | 7 | 1440 | < th | dose independent | rect. | 23 | 2 |
| 504 | 218.43 | wind | 9 | 1440 | < th | dose independent | rect. | 21 | 8 |
| 520 | 218.61 | humidity | 60 | 90 | < th | dose independent | rect. | 9 | 1 |
| 548 | 218.75 | wind | 7 | 1440 | < th | dose independent | rect. | 21 | 5 |
| 574 | 218.81 | wind | 9 | 720 | > th | dose independent | rect. | 19 | 3 |
| 575 | 218.82 | wind | 5 | 720 | < th | dose independent | rect. | 20 | 12 |
| 576 | 218.84 | wind | 9 | 720 | > th | dose dependent | rect. | 19 | 2 |
| 669 | 219.15 | wind | 5 | 720 | < th | dose independent | rect. | 23 | 9 |