Os02g0754400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Myb, DNA-binding domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0754400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Myb, DNA-binding domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
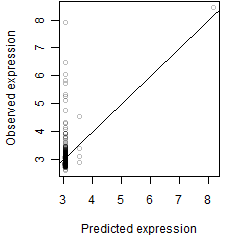
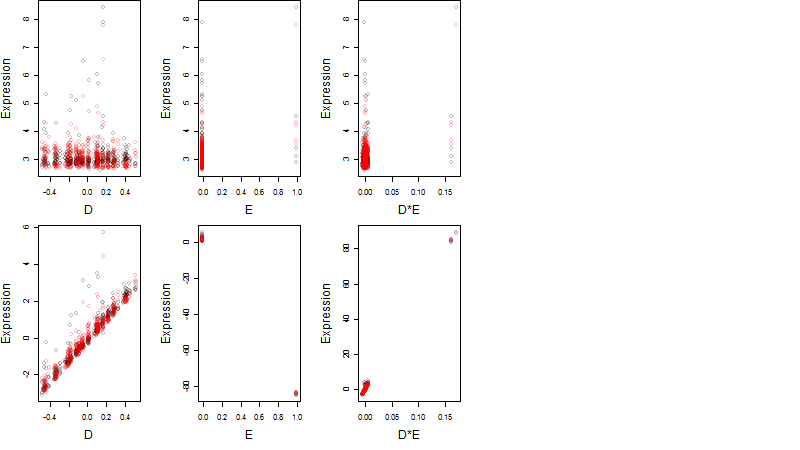
Dependence on each variable
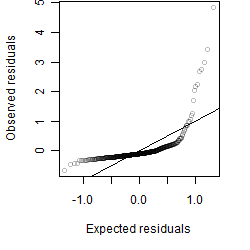
Residual plot
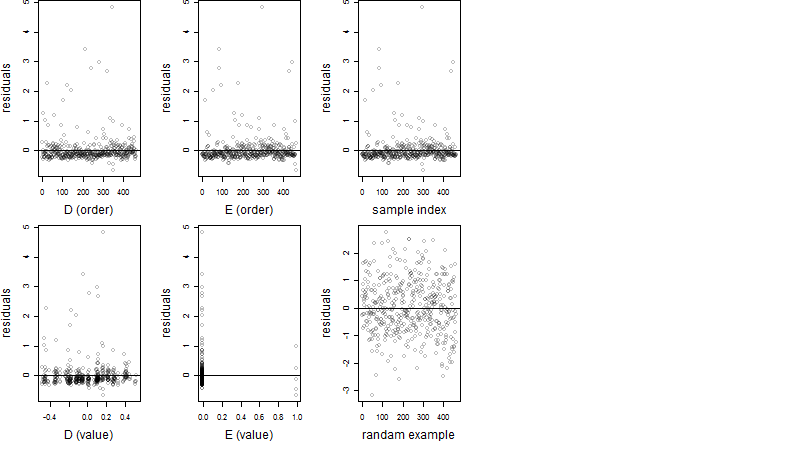
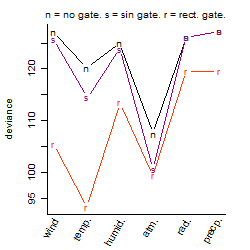
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 101.36 | 0.473 | 2.07 | 6.1 | 0.0886 | -84.3 | -0.258 | 518 | -0.00246 | 6.36 | 994.8 | 669 | -- | -- |
| 128.62 | 0.532 | 3.08 | 0.0134 | 0.106 | 0.0174 | -0.208 | -- | -0.0208 | 5.7 | 994.8 | 669 | -- | -- |
| 99.49 | 0.465 | 2.14 | 5.64 | 0.0983 | -84.7 | -- | 522 | -0.00346 | 7.55 | 994.5 | 691 | -- | -- |
| 119.43 | 0.512 | 3.08 | -0.065 | 0.11 | 1.11 | -- | -- | 0.00987 | 5.59 | 999.4 | 704 | -- | -- |
| 100 | 0.466 | 2.14 | 5.65 | -- | -85 | -- | 523 | -0.00238 | -- | 994.6 | 685 | -- | -- |
| 128.62 | 0.532 | 3.08 | 0.0135 | 0.106 | -- | -0.208 | -- | -0.0208 | 5.7 | -- | -- | -- | -- |
| 128.72 | 0.531 | 3.08 | 0.0123 | 0.104 | -- | -- | -- | -0.02 | 5.66 | -- | -- | -- | -- |
| 120.04 | 0.513 | 3.08 | -0.0608 | -- | 1.12 | -- | -- | 0.0123 | -- | 999.4 | 709 | -- | -- |
| 119.5 | 0.512 | 3.08 | -- | 0.11 | 1.11 | -- | -- | 0.0163 | 5.62 | 999.3 | 704 | -- | -- |
| 129.29 | 0.531 | 3.09 | 0.0156 | -- | -- | -- | -- | -0.0175 | -- | -- | -- | -- | -- |
| 128.73 | 0.531 | 3.08 | -- | 0.105 | -- | -- | -- | -0.0214 | 5.65 | -- | -- | -- | -- |
| 120.11 | 0.512 | 3.08 | -- | -- | 1.11 | -- | -- | 0.0183 | -- | 999.3 | 709 | -- | -- |
| 129.3 | 0.531 | 3.09 | -- | -- | -- | -- | -- | -0.0192 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 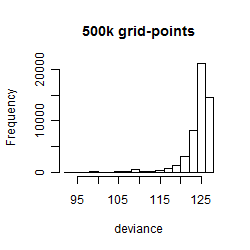
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 93.36 | temperature | 25 | 90 | > th | dose independent | rect. | 1 | 1 |
| 2 | 97.27 | temperature | 25 | 90 | > th | dose dependent | rect. | 1 | 1 |
| 4 | 99.46 | atmosphere | 995 | 720 | < th | dose independent | rect. | 14 | 1 |
| 45 | 99.46 | atmosphere | 995 | 720 | < th | dose dependent | rect. | 14 | 1 |
| 86 | 99.97 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 1 | 1 |
| 93 | 99.97 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 1 | 1 |
| 103 | 100.08 | temperature | 25 | 10 | > th | dose dependent | rect. | 1 | 1 |
| 119 | 100.64 | atmosphere | 995 | 720 | > th | dose independent | sin | 7 | NA |
| 121 | 101.02 | atmosphere | 995 | 720 | < th | dose independent | sin | 6 | NA |
| 125 | 101.40 | atmosphere | 995 | 720 | > th | dose independent | sin | 5 | NA |
| 130 | 102.09 | atmosphere | 995 | 720 | < th | dose dependent | sin | 7 | NA |
| 167 | 105.14 | atmosphere | 995 | 720 | > th | dose independent | sin | 1 | NA |
| 168 | 105.38 | atmosphere | 1000 | 30 | < th | dose dependent | rect. | 11 | 1 |
| 177 | 105.43 | wind | 1 | 10 | < th | dose independent | rect. | 12 | 1 |
| 179 | 105.43 | wind | 1 | 10 | < th | dose dependent | rect. | 12 | 1 |
| 226 | 107.33 | atmosphere | 1000 | 90 | < th | dose independent | rect. | 10 | 1 |
| 230 | 107.38 | temperature | 30 | 90 | > th | dose dependent | rect. | 10 | 1 |
| 231 | 107.40 | atmosphere | 995 | 720 | > th | dose independent | no | NA | NA |
| 255 | 107.62 | atmosphere | 1000 | 10 | < th | dose independent | rect. | 11 | 1 |
| 277 | 108.40 | atmosphere | 995 | 720 | < th | dose independent | no | NA | NA |
| 500 | 109.20 | atmosphere | 995 | 720 | < th | dose dependent | no | NA | NA |
| 693 | 109.30 | atmosphere | 995 | 720 | > th | dose independent | sin | 15 | NA |
| 800 | 111.80 | temperature | 30 | 90 | > th | dose independent | rect. | 10 | 1 |
| 840 | 113.28 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 1 | 12 |
| 851 | 113.48 | humidity | 90 | 90 | < th | dose independent | rect. | 1 | 1 |
| 915 | 114.43 | temperature | 30 | 90 | > th | dose dependent | sin | 9 | NA |
| 998 | 114.61 | atmosphere | 1000 | 10 | < th | dose dependent | rect. | 19 | 18 |