Os02g0752300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Conserved hypothetical protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0752300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Conserved hypothetical protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

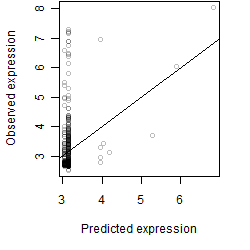 __
__
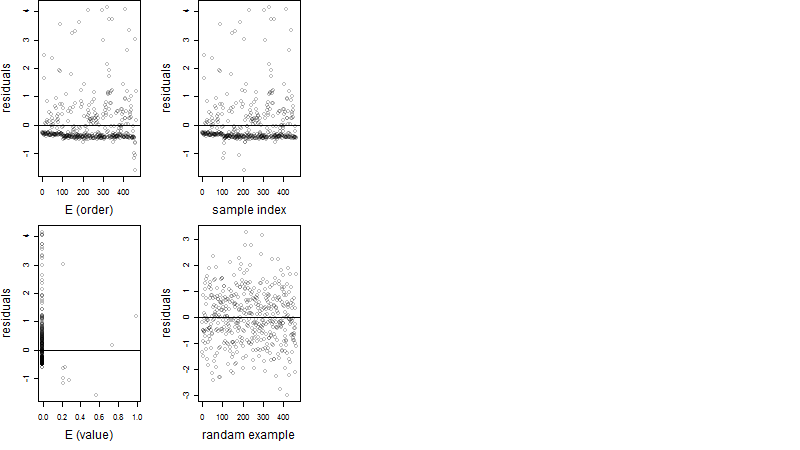
Dependence on each variable

Residual plot
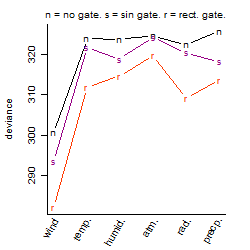
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 284.45 | 0.792 | 3.21 | 0.602 | 0.248 | 8.11 | -0.329 | 17.7 | -0.0425 | 3.25 | 10.34 | 126 | -- | -- |
| 300.4 | 0.807 | 3.17 | 0.478 | 0.207 | 4.1 | -0.437 | -- | -0.00169 | 23.7 | 9.4 | 54 | -- | -- |
| 284.69 | 0.786 | 3.21 | 0.585 | 0.248 | 8.14 | -- | 18.5 | -0.0529 | 3.56 | 10.36 | 125 | -- | -- |
| 298.7 | 0.805 | 3.18 | 0.433 | 0.218 | 4.43 | -- | -- | -0.0133 | 2.65 | 9.545 | 49 | -- | -- |
| 287.76 | 0.79 | 3.23 | 0.69 | -- | 10.7 | -- | 30 | -0.0393 | -- | 10.39 | 116 | -- | -- |
| 332.16 | 0.854 | 3.18 | 0.297 | 0.0745 | -- | 0.738 | -- | -0.0927 | 6.93 | -- | -- | -- | -- |
| 332.73 | 0.854 | 3.18 | 0.306 | 0.143 | -- | -- | -- | -0.0969 | 3.28 | -- | -- | -- | -- |
| 300.94 | 0.808 | 3.17 | 0.486 | -- | 3.79 | -- | -- | -0.0313 | -- | 9.532 | 49 | -- | -- |
| 304.08 | 0.812 | 3.18 | -- | 0.213 | 4.07 | -- | -- | -0.0842 | 2.19 | 9.75 | 49 | -- | -- |
| 333.88 | 0.854 | 3.18 | 0.312 | -- | -- | -- | -- | -0.0931 | -- | -- | -- | -- | -- |
| 335.2 | 0.856 | 3.19 | -- | 0.149 | -- | -- | -- | -0.131 | 3.28 | -- | -- | -- | -- |
| 306.89 | 0.816 | 3.18 | -- | -- | 3.69 | -- | -- | -0.0893 | -- | 9.687 | 49 | -- | -- |
| 336.45 | 0.856 | 3.19 | -- | -- | -- | -- | -- | -0.128 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 282.06 | wind | 7 | 270 | > th | dose dependent | rect. | 13 | 1 |
| 3 | 282.76 | wind | 9 | 270 | > th | dose independent | rect. | 13 | 1 |
| 4 | 282.96 | wind | 9 | 270 | > th | dose independent | rect. | 12 | 3 |
| 5 | 283.34 | wind | 9 | 270 | > th | dose dependent | rect. | 12 | 3 |
| 7 | 283.72 | wind | 9 | 270 | > th | dose dependent | rect. | 11 | 5 |
| 9 | 283.87 | wind | 9 | 90 | > th | dose independent | rect. | 12 | 5 |
| 35 | 288.33 | wind | 9 | 270 | > th | dose dependent | rect. | 1 | 15 |
| 52 | 289.87 | wind | 9 | 270 | > th | dose independent | rect. | 1 | 15 |
| 64 | 290.20 | wind | 9 | 90 | > th | dose dependent | rect. | 12 | 9 |
| 65 | 290.29 | wind | 9 | 90 | > th | dose dependent | rect. | 12 | 7 |
| 75 | 290.87 | wind | 9 | 90 | > th | dose dependent | rect. | 12 | 14 |
| 91 | 291.22 | wind | 9 | 90 | > th | dose independent | rect. | 12 | 14 |
| 197 | 293.56 | wind | 9 | 270 | > th | dose dependent | sin | 2 | NA |
| 235 | 294.45 | wind | 9 | 270 | < th | dose independent | sin | 2 | NA |
| 310 | 295.26 | wind | 9 | 270 | > th | dose independent | sin | 2 | NA |
| 426 | 297.51 | wind | 9 | 90 | > th | dose dependent | sin | 19 | NA |
| 493 | 299.19 | wind | 9 | 30 | < th | dose independent | sin | 17 | NA |
| 676 | 300.51 | wind | 9 | 90 | > th | dose independent | sin | 19 | NA |
| 698 | 300.59 | wind | 9 | 270 | > th | dose dependent | no | NA | NA |
| 877 | 301.81 | wind | 9 | 270 | > th | dose independent | no | NA | NA |
| 912 | 301.87 | wind | 9 | 270 | > th | dose dependent | rect. | 15 | 23 |
| 917 | 301.90 | wind | 9 | 270 | < th | dose independent | no | NA | NA |