Os02g0697400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1) (4-coumaroyl-CoA synthase 1) (Clone 4CL14) (Fragment).

FiT-DB / Search/ Help/ Sample detail
|
Os02g0697400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to 4-coumarate--CoA ligase 1 (EC 6.2.1.12) (4CL 1) (4-coumaroyl-CoA synthase 1) (Clone 4CL14) (Fragment).
|
|
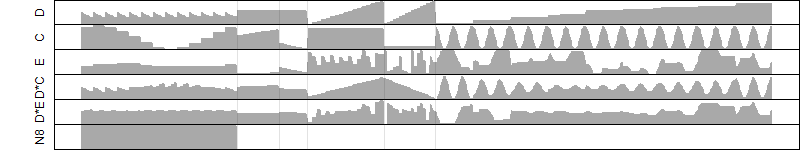
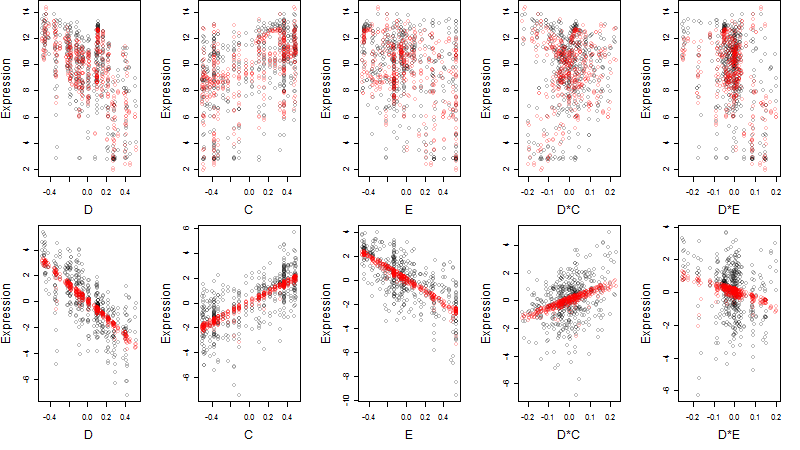
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable
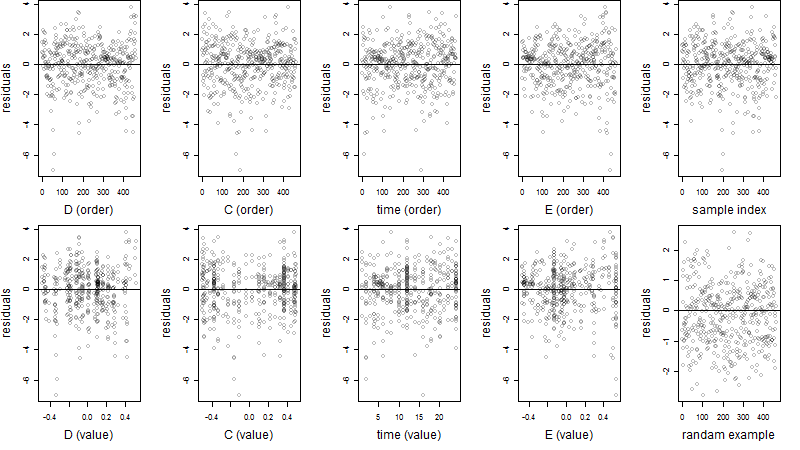
Residual plot
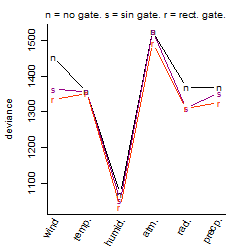
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 987.7 | 1.48 | 9.7 | -6.5 | 3.95 | -4.83 | 5.82 | -4.16 | -1.13 | 9.24 | 73 | 1256 | -- | -- |
| 1028.39 | 1.49 | 9.76 | -6.92 | 4.03 | -4.82 | 5.95 | -- | -1.19 | 9.16 | 72 | 1247 | -- | -- |
| 1093.75 | 1.54 | 9.71 | -6.17 | 3.65 | -4.89 | -- | -3.56 | -1.13 | 8.92 | 75 | 1317 | -- | -- |
| 1126.37 | 1.56 | 9.75 | -6.89 | 3.99 | -4.8 | -- | -- | -1.22 | 9.14 | 73 | 1246 | -- | -- |
| 1761.47 | 1.95 | 9.81 | -6.14 | -- | -4.72 | -- | -4.18 | -1.05 | -- | 74.87 | 1430 | -- | -- |
| 1855.28 | 2.02 | 9.63 | -6.15 | 3.53 | -- | 4.95 | -- | -0.632 | 8.26 | -- | -- | -- | -- |
| 1930.77 | 2.06 | 9.64 | -6 | 3.53 | -- | -- | -- | -0.637 | 8.18 | -- | -- | -- | -- |
| 1801.33 | 1.98 | 9.88 | -6.73 | -- | -4.96 | -- | -- | -1.25 | -- | 74.57 | 1420 | -- | -- |
| 2117.48 | 2.14 | 9.6 | -- | 5.14 | -6.34 | -- | -- | -0.54 | 8.15 | 84 | 2148 | -- | -- |
| 2616.65 | 2.39 | 9.73 | -5.98 | -- | -- | -- | -- | -0.596 | -- | -- | -- | -- | -- |
| 2881.04 | 2.51 | 9.49 | -- | 3.5 | -- | -- | -- | 0.0322 | 8.31 | -- | -- | -- | -- |
| 2798.92 | 2.46 | 9.78 | -- | -- | -7.3 | -- | -- | -0.818 | -- | 85.99 | 1487 | -- | -- |
| 3558.98 | 2.78 | 9.58 | -- | -- | -- | -- | -- | 0.0676 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance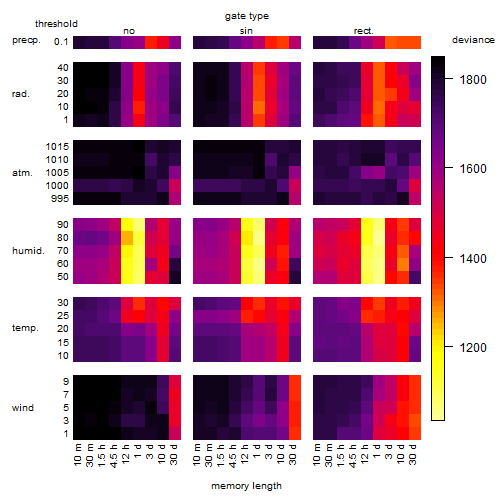
|
Histogram 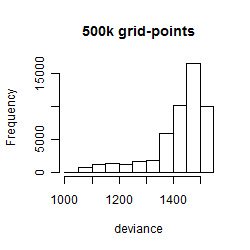
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1033.58 | humidity | 80 | 1440 | < th | dose independent | rect. | 7 | 8 |
| 6 | 1050.65 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 9 |
| 8 | 1051.28 | humidity | 60 | 1440 | > th | dose dependent | rect. | 6 | 16 |
| 12 | 1052.67 | humidity | 80 | 1440 | < th | dose independent | sin | 1 | NA |
| 24 | 1055.89 | humidity | 80 | 1440 | < th | dose independent | rect. | 6 | 14 |
| 33 | 1056.68 | humidity | 60 | 1440 | > th | dose dependent | sin | 0 | NA |
| 40 | 1058.58 | humidity | 80 | 1440 | > th | dose independent | sin | 1 | NA |
| 80 | 1063.24 | humidity | 90 | 1440 | < th | dose dependent | rect. | 16 | 22 |
| 85 | 1063.56 | humidity | 80 | 1440 | > th | dose independent | rect. | 6 | 14 |
| 102 | 1064.87 | humidity | 90 | 1440 | < th | dose dependent | rect. | 2 | 21 |
| 167 | 1068.52 | humidity | 90 | 1440 | < th | dose dependent | rect. | 13 | 23 |
| 175 | 1068.80 | humidity | 70 | 1440 | > th | dose independent | rect. | 18 | 20 |
| 210 | 1070.41 | humidity | 90 | 1440 | < th | dose dependent | no | NA | NA |
| 221 | 1070.84 | humidity | 90 | 1440 | < th | dose dependent | rect. | 20 | 23 |
| 327 | 1077.00 | humidity | 50 | 1440 | > th | dose dependent | no | NA | NA |
| 339 | 1077.65 | humidity | 90 | 1440 | < th | dose dependent | rect. | 11 | 23 |
| 349 | 1078.32 | humidity | 50 | 1440 | > th | dose dependent | rect. | 16 | 23 |
| 359 | 1078.85 | humidity | 50 | 1440 | > th | dose dependent | rect. | 18 | 23 |
| 368 | 1079.36 | humidity | 90 | 1440 | < th | dose dependent | sin | 1 | NA |
| 379 | 1079.75 | humidity | 50 | 1440 | > th | dose dependent | rect. | 13 | 23 |
| 418 | 1082.71 | humidity | 70 | 1440 | > th | dose independent | rect. | 15 | 23 |
| 474 | 1086.30 | humidity | 50 | 1440 | > th | dose dependent | rect. | 11 | 23 |
| 529 | 1088.07 | humidity | 70 | 1440 | > th | dose independent | rect. | 6 | 23 |
| 531 | 1088.08 | humidity | 70 | 1440 | > th | dose independent | rect. | 6 | 21 |
| 543 | 1088.12 | humidity | 70 | 1440 | > th | dose independent | rect. | 4 | 23 |
| 546 | 1088.14 | humidity | 70 | 1440 | > th | dose independent | no | NA | NA |
| 685 | 1095.60 | humidity | 70 | 1440 | > th | dose independent | rect. | 11 | 23 |
| 851 | 1105.54 | humidity | 60 | 1440 | > th | dose dependent | rect. | 11 | 2 |
| 912 | 1109.66 | humidity | 70 | 1440 | < th | dose independent | rect. | 17 | 23 |
| 926 | 1110.26 | humidity | 70 | 1440 | < th | dose independent | rect. | 14 | 23 |