Os02g0484200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Transferase family protein.

FiT-DB / Search/ Help/ Sample detail
|
Os02g0484200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Transferase family protein.
|
|
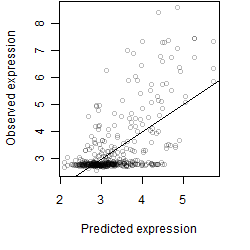
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__

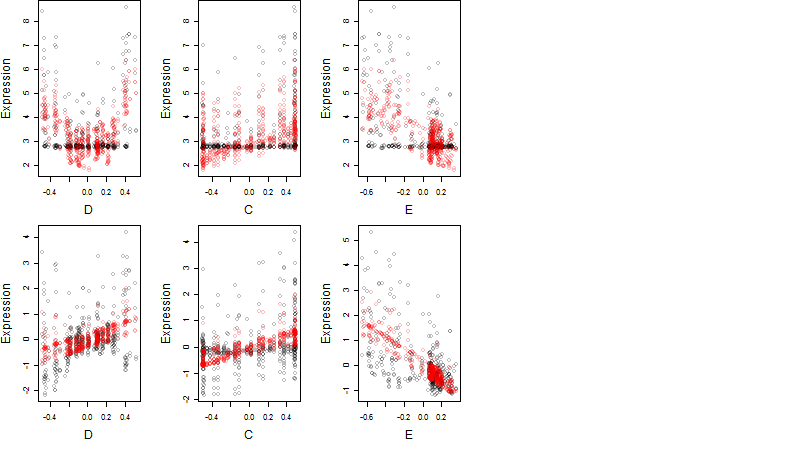
Dependence on each variable
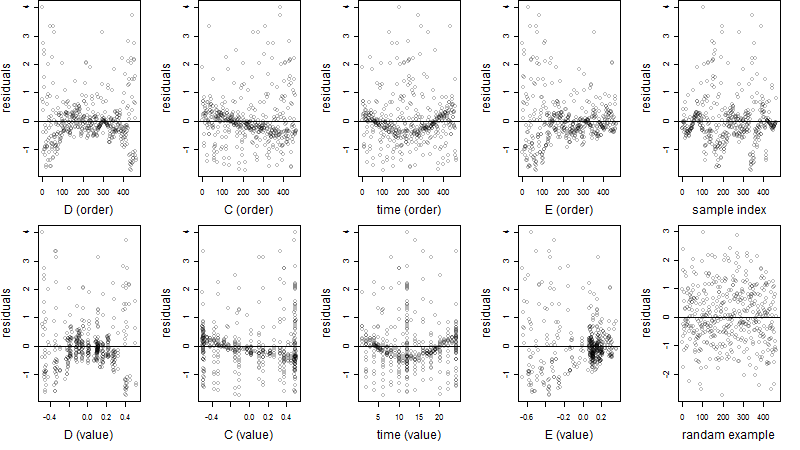
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 310.31 | 0.828 | 3.3 | 0.768 | 0.746 | -2.25 | 1.91 | -1.12 | -0.0554 | 11.6 | 1010 | 24475 | 12.5 | -- |
| 310.95 | 0.821 | 3.28 | 1.08 | 0.698 | -2.38 | 1.8 | -- | -0.0396 | 12 | 1009 | 24603 | 13.7 | -- |
| 324.3 | 0.839 | 3.31 | 0.869 | 0.722 | -2.35 | -- | -1.11 | -0.0339 | 12.3 | 1010 | 24518 | 11.7 | -- |
| 323.63 | 0.838 | 3.27 | 1.2 | 0.79 | -2.38 | -- | -- | -0.0137 | 12.8 | 1009 | 24805 | 16.2 | -- |
| 343.44 | 0.863 | 3.33 | 0.635 | -- | -2.62 | -- | -2.1 | -0.193 | -- | 1009 | 22479 | 13.1 | -- |
| 445.23 | 0.989 | 3.36 | 0.017 | 0.805 | -- | 2 | -- | -0.436 | 11.8 | -- | -- | -- | -- |
| 461.2 | 1.01 | 3.36 | 0.102 | 0.811 | -- | -- | -- | -0.423 | 12.6 | -- | -- | -- | -- |
| 350.29 | 0.872 | 3.29 | 1.02 | -- | -2.72 | -- | -- | -0.0535 | -- | 1009 | 22393 | 12.7 | -- |
| 340.11 | 0.859 | 3.34 | -- | 0.743 | -1.93 | -- | -- | -0.192 | 13.4 | 1011 | 20898 | 17.5 | -- |
| 502.44 | 1.05 | 3.37 | 0.0732 | -- | -- | -- | -- | -0.437 | -- | -- | -- | -- | -- |
| 461.47 | 1 | 3.36 | -- | 0.81 | -- | -- | -- | -0.434 | 12.6 | -- | -- | -- | -- |
| 365.91 | 0.891 | 3.3 | -- | -- | -1.92 | -- | -- | -0.168 | -- | 1011 | 21137 | 16.2 | -- |
| 502.58 | 1.05 | 3.38 | -- | -- | -- | -- | -- | -0.445 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance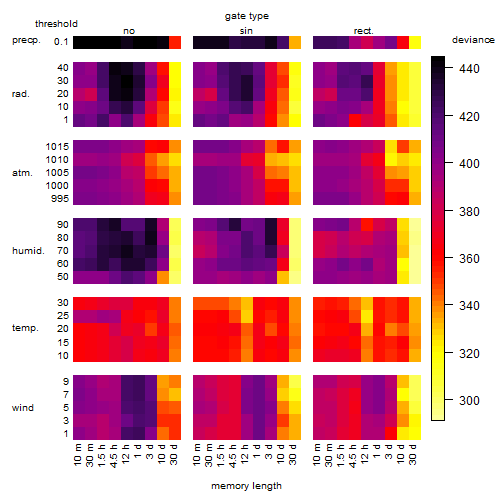
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 291.08 | humidity | 50 | 43200 | > th | dose dependent | rect. | 16 | 7 |
| 6 | 291.66 | humidity | 90 | 43200 | < th | dose dependent | rect. | 16 | 6 |
| 12 | 292.12 | humidity | 70 | 43200 | > th | dose dependent | rect. | 20 | 1 |
| 47 | 293.20 | humidity | 90 | 43200 | < th | dose dependent | rect. | 16 | 1 |
| 84 | 294.39 | humidity | 80 | 43200 | > th | dose independent | rect. | 16 | 8 |
| 91 | 294.63 | humidity | 80 | 43200 | < th | dose independent | rect. | 16 | 8 |
| 101 | 294.76 | humidity | 50 | 43200 | > th | dose dependent | sin | 18 | NA |
| 160 | 296.04 | humidity | 90 | 43200 | < th | dose dependent | sin | 19 | NA |
| 232 | 296.95 | humidity | 70 | 43200 | < th | dose independent | rect. | 14 | 8 |
| 244 | 297.03 | humidity | 70 | 43200 | > th | dose independent | rect. | 14 | 8 |
| 247 | 297.06 | humidity | 70 | 43200 | < th | dose independent | rect. | 13 | 10 |
| 263 | 297.20 | humidity | 70 | 43200 | > th | dose independent | rect. | 13 | 10 |
| 322 | 297.67 | humidity | 70 | 43200 | < th | dose independent | sin | 18 | NA |
| 325 | 297.71 | humidity | 60 | 43200 | > th | dose independent | rect. | 12 | 1 |
| 335 | 297.77 | humidity | 80 | 43200 | < th | dose independent | rect. | 18 | 5 |
| 347 | 297.86 | humidity | 70 | 43200 | > th | dose independent | sin | 19 | NA |
| 358 | 297.92 | humidity | 70 | 43200 | < th | dose independent | rect. | 8 | 16 |
| 360 | 297.96 | humidity | 70 | 43200 | > th | dose independent | rect. | 6 | 17 |
| 611 | 299.14 | humidity | 60 | 43200 | < th | dose independent | rect. | 12 | 2 |
| 778 | 299.91 | humidity | 80 | 43200 | < th | dose independent | rect. | 20 | 1 |
| 799 | 299.98 | humidity | 80 | 43200 | > th | dose independent | rect. | 20 | 2 |
| 823 | 300.05 | humidity | 80 | 43200 | < th | dose independent | rect. | 19 | 3 |
| 877 | 300.28 | humidity | 70 | 43200 | < th | dose independent | no | NA | NA |
| 878 | 300.28 | humidity | 70 | 43200 | < th | dose independent | rect. | 11 | 23 |
| 900 | 300.36 | humidity | 70 | 43200 | > th | dose independent | rect. | 12 | 23 |
| 933 | 300.50 | humidity | 70 | 43200 | > th | dose independent | no | NA | NA |
| 986 | 300.67 | humidity | 50 | 43200 | > th | dose dependent | no | NA | NA |