Os02g0435900
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, unclassifiable transcript.
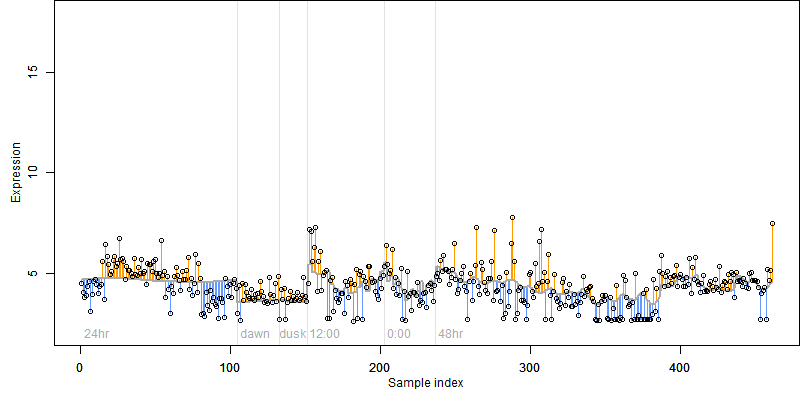

FiT-DB / Search/ Help/ Sample detail
|
Os02g0435900 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, unclassifiable transcript.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

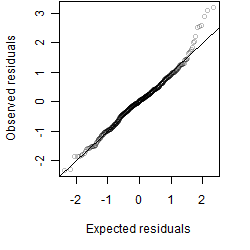
Residual plot

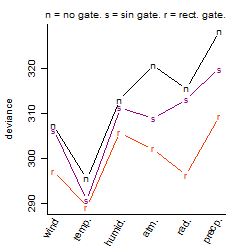
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 295.05 | 0.807 | 4.17 | -0.0368 | 0.822 | -2.29 | -1.62 | 0.706 | 0.702 | 17 | -7.442 | 647 | -- | -- |
| 295.4 | 0.8 | 4.19 | -0.162 | 0.846 | -2.29 | -1.5 | -- | 0.669 | 16.9 | 10.46 | 629 | -- | -- |
| 300.42 | 0.807 | 4.19 | -0.181 | 0.979 | -2.37 | -- | -0.025 | 0.668 | 16.4 | -24.05 | 513 | -- | -- |
| 300.36 | 0.807 | 4.19 | -0.183 | 1.01 | -2.36 | -- | -- | 0.666 | 16.3 | -25.6 | 478 | -- | -- |
| 312.41 | 0.823 | 4.18 | 0.0624 | -- | -1.96 | -- | 0.834 | 0.674 | -- | -61.85 | 1128 | -- | -- |
| 362.19 | 0.892 | 4.25 | -0.695 | 0.593 | -- | -0.87 | -- | 0.321 | 14.7 | -- | -- | -- | -- |
| 363.95 | 0.893 | 4.25 | -0.717 | 0.593 | -- | -- | -- | 0.314 | 13.8 | -- | -- | -- | -- |
| 313.01 | 0.824 | 4.18 | -0.129 | -- | -2.07 | -- | -- | 0.66 | -- | -55.08 | 1092 | -- | -- |
| 301.15 | 0.808 | 4.18 | -- | 1.03 | -2.44 | -- | -- | 0.696 | 16.3 | -8.667 | 488 | -- | -- |
| 385.22 | 0.917 | 4.26 | -0.742 | -- | -- | -- | -- | 0.301 | -- | -- | -- | -- | -- |
| 377.51 | 0.909 | 4.24 | -- | 0.606 | -- | -- | -- | 0.394 | 13.9 | -- | -- | -- | -- |
| 313.38 | 0.824 | 4.17 | -- | -- | -2.13 | -- | -- | 0.683 | -- | -7.283 | 1095 | -- | -- |
| 399.74 | 0.933 | 4.24 | -- | -- | -- | -- | -- | 0.383 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance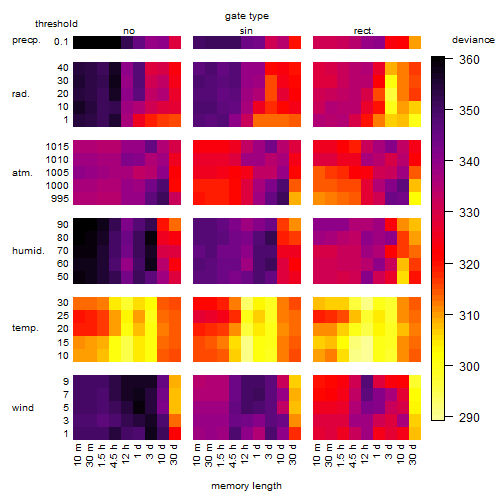
|
Histogram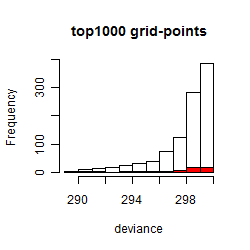 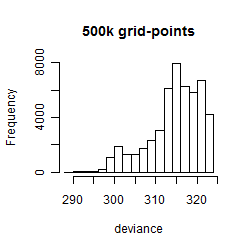
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 289.19 | temperature | 10 | 720 | > th | dose dependent | rect. | 15 | 17 |
| 8 | 290.66 | temperature | 15 | 720 | > th | dose dependent | sin | 12 | NA |
| 11 | 290.96 | temperature | 10 | 720 | > th | dose dependent | rect. | 10 | 22 |
| 17 | 291.38 | temperature | 30 | 720 | < th | dose dependent | rect. | 16 | 17 |
| 46 | 293.23 | temperature | 30 | 720 | < th | dose dependent | sin | 12 | NA |
| 82 | 294.50 | temperature | 10 | 720 | > th | dose dependent | rect. | 6 | 23 |
| 88 | 294.83 | temperature | 15 | 720 | > th | dose dependent | rect. | 15 | 23 |
| 90 | 294.85 | temperature | 10 | 720 | > th | dose dependent | rect. | 4 | 23 |
| 98 | 295.12 | temperature | 10 | 720 | > th | dose dependent | rect. | 19 | 23 |
| 104 | 295.36 | temperature | 10 | 720 | > th | dose dependent | rect. | 24 | 23 |
| 110 | 295.50 | temperature | 15 | 720 | > th | dose dependent | rect. | 19 | 10 |
| 113 | 295.55 | temperature | 10 | 720 | > th | dose dependent | no | NA | NA |
| 135 | 296.01 | temperature | 10 | 720 | > th | dose dependent | rect. | 2 | 23 |
| 146 | 296.24 | temperature | 15 | 720 | > th | dose dependent | rect. | 17 | 23 |
| 154 | 296.34 | radiation | 20 | 4320 | < th | dose independent | rect. | 16 | 1 |
| 207 | 296.99 | temperature | 30 | 4320 | < th | dose dependent | rect. | 2 | 1 |
| 212 | 297.07 | temperature | 10 | 4320 | > th | dose dependent | rect. | 2 | 1 |
| 213 | 297.08 | temperature | 30 | 720 | < th | dose dependent | rect. | 7 | 23 |
| 222 | 297.17 | temperature | 25 | 4320 | < th | dose independent | rect. | 13 | 20 |
| 228 | 297.24 | wind | 9 | 43200 | > th | dose dependent | rect. | 19 | 12 |
| 252 | 297.46 | temperature | 25 | 4320 | > th | dose independent | rect. | 13 | 20 |
| 306 | 297.85 | radiation | 20 | 4320 | < th | dose independent | rect. | 16 | 14 |
| 317 | 297.92 | temperature | 30 | 720 | < th | dose dependent | no | NA | NA |
| 341 | 298.05 | radiation | 20 | 4320 | < th | dose independent | rect. | 16 | 4 |
| 387 | 298.30 | temperature | 30 | 720 | < th | dose dependent | rect. | 21 | 23 |
| 413 | 298.41 | temperature | 25 | 4320 | < th | dose independent | sin | 14 | NA |
| 435 | 298.53 | temperature | 10 | 4320 | > th | dose dependent | rect. | 21 | 12 |
| 437 | 298.54 | temperature | 25 | 4320 | > th | dose dependent | rect. | 13 | 21 |
| 441 | 298.56 | temperature | 10 | 4320 | > th | dose dependent | rect. | 24 | 9 |
| 442 | 298.56 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 12 |
| 453 | 298.59 | temperature | 25 | 4320 | < th | dose independent | rect. | 19 | 20 |
| 461 | 298.60 | temperature | 30 | 4320 | < th | dose dependent | rect. | 24 | 9 |
| 467 | 298.63 | temperature | 25 | 4320 | > th | dose dependent | rect. | 15 | 19 |
| 480 | 298.69 | temperature | 25 | 4320 | > th | dose dependent | sin | 7 | NA |
| 486 | 298.70 | temperature | 25 | 4320 | > th | dose independent | sin | 14 | NA |
| 490 | 298.72 | radiation | 20 | 4320 | < th | dose independent | rect. | 16 | 11 |
| 532 | 298.82 | temperature | 25 | 4320 | > th | dose dependent | rect. | 19 | 19 |
| 536 | 298.83 | temperature | 30 | 720 | < th | dose dependent | rect. | 5 | 23 |
| 548 | 298.86 | temperature | 25 | 4320 | > th | dose dependent | rect. | 7 | 2 |
| 559 | 298.88 | temperature | 10 | 4320 | > th | dose dependent | sin | 10 | NA |
| 572 | 298.90 | temperature | 30 | 720 | < th | dose dependent | rect. | 1 | 23 |
| 634 | 299.06 | temperature | 20 | 4320 | > th | dose dependent | rect. | 17 | 10 |
| 651 | 299.10 | temperature | 30 | 4320 | < th | dose dependent | sin | 11 | NA |
| 654 | 299.11 | temperature | 25 | 4320 | > th | dose independent | rect. | 14 | 13 |
| 676 | 299.14 | temperature | 25 | 4320 | > th | dose independent | rect. | 19 | 20 |
| 758 | 299.27 | temperature | 25 | 4320 | > th | dose dependent | rect. | 6 | 20 |
| 786 | 299.31 | temperature | 30 | 720 | < th | dose dependent | rect. | 3 | 23 |
| 797 | 299.33 | temperature | 20 | 4320 | > th | dose dependent | rect. | 13 | 15 |
| 815 | 299.34 | temperature | 25 | 4320 | > th | dose dependent | rect. | 23 | 23 |
| 817 | 299.35 | wind | 9 | 43200 | > th | dose dependent | rect. | 19 | 5 |
| 820 | 299.36 | temperature | 20 | 4320 | > th | dose dependent | sin | 14 | NA |
| 829 | 299.37 | temperature | 25 | 4320 | > th | dose dependent | no | NA | NA |
| 850 | 299.39 | temperature | 10 | 4320 | > th | dose dependent | rect. | 13 | 15 |
| 863 | 299.41 | temperature | 10 | 1440 | > th | dose dependent | rect. | 6 | 3 |
| 920 | 299.47 | temperature | 10 | 4320 | > th | dose dependent | rect. | 13 | 20 |
| 949 | 299.52 | temperature | 25 | 4320 | > th | dose dependent | rect. | 6 | 16 |