Os02g0327500
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein of unknown function DUF266, plant family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0327500 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein of unknown function DUF266, plant family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

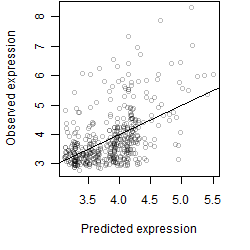
Dependence on each variable

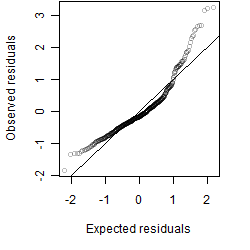
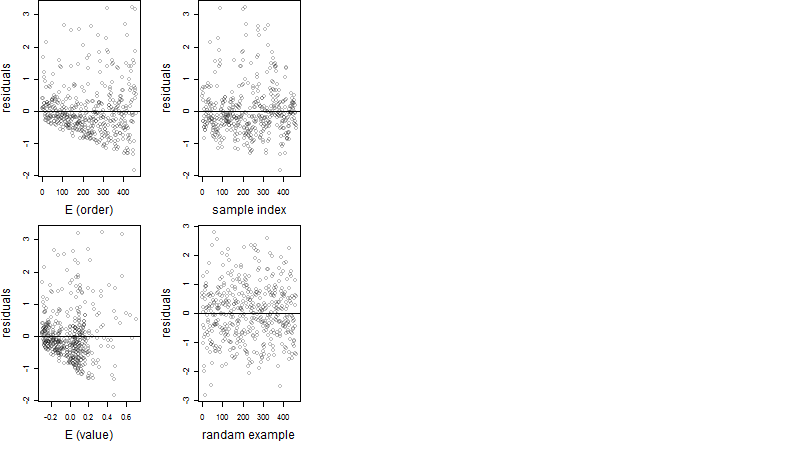
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = < th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 250.78 | 0.744 | 3.85 | 0.54 | 3.34 | 5.46 | 1.15 | 0.737 | 0.143 | 15.8 | 9.23 | 750 | 11.2 | -- |
| 250.73 | 0.737 | 3.85 | 0.561 | 3.31 | 5.48 | 0.555 | -- | 0.148 | 15.8 | 9.111 | 756 | 11.4 | -- |
| 251.8 | 0.739 | 3.83 | 0.645 | 3.24 | 5.54 | -- | -0.181 | 0.182 | 15.5 | 9.255 | 753 | 11.7 | -- |
| 251.49 | 0.739 | 3.84 | 0.595 | 3.15 | 5.39 | -- | -- | 0.159 | 15.7 | 8.863 | 762 | 11.6 | -- |
| 296.42 | 0.802 | 3.75 | 0.531 | -- | 1.67 | -- | 1.6 | 0.301 | -- | 46.45 | 2187 | 12.5 | -- |
| 309.53 | 0.825 | 3.85 | 0.536 | 0.865 | -- | 0.001 | -- | 0.0227 | 1.79 | -- | -- | -- | -- |
| 309.53 | 0.824 | 3.85 | 0.536 | 0.865 | -- | -- | -- | 0.0227 | 1.79 | -- | -- | -- | -- |
| 260.69 | 0.752 | 3.83 | 0.615 | -- | 2.02 | -- | -- | 0.234 | -- | 12.98 | 854 | 15.5 | -- |
| 259.74 | 0.751 | 3.86 | -- | 2.94 | 5.13 | -- | -- | 0.0942 | 15.8 | 10 | 767 | 11.6 | -- |
| 354.9 | 0.88 | 3.84 | 0.571 | -- | -- | -- | -- | 0.0423 | -- | -- | -- | -- | -- |
| 317.1 | 0.833 | 3.87 | -- | 0.875 | -- | -- | -- | -0.037 | 1.81 | -- | -- | -- | -- |
| 268.46 | 0.763 | 3.83 | -- | -- | 2.38 | -- | -- | 0.101 | -- | 9.91 | 844 | 15.9 | -- |
| 363.51 | 0.89 | 3.86 | -- | -- | -- | -- | -- | -0.0211 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram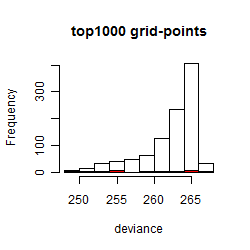 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 248.60 | radiation | 10 | 720 | < th | dose dependent | rect. | 14 | 4 |
| 8 | 250.08 | radiation | 1 | 720 | < th | dose independent | rect. | 8 | 10 |
| 14 | 250.73 | radiation | 1 | 720 | < th | dose independent | rect. | 9 | 18 |
| 26 | 252.27 | radiation | 1 | 720 | < th | dose independent | sin | 19 | NA |
| 29 | 252.47 | radiation | 10 | 720 | < th | dose dependent | rect. | 14 | 11 |
| 47 | 253.76 | radiation | 10 | 720 | < th | dose dependent | rect. | 13 | 14 |
| 50 | 253.89 | radiation | 10 | 720 | < th | dose dependent | sin | 17 | NA |
| 62 | 254.50 | radiation | 1 | 720 | > th | dose independent | rect. | 9 | 10 |
| 75 | 254.85 | radiation | 10 | 720 | < th | dose dependent | rect. | 14 | 9 |
| 84 | 255.51 | radiation | 1 | 720 | > th | dose independent | rect. | 8 | 22 |
| 85 | 255.52 | radiation | 1 | 720 | > th | dose independent | sin | 20 | NA |
| 88 | 255.73 | radiation | 10 | 720 | < th | dose dependent | rect. | 10 | 16 |
| 192 | 259.59 | humidity | 50 | 720 | > th | dose dependent | sin | 16 | NA |
| 220 | 260.40 | humidity | 50 | 720 | > th | dose dependent | rect. | 13 | 14 |
| 374 | 262.67 | humidity | 80 | 720 | > th | dose independent | rect. | 11 | 11 |
| 594 | 264.19 | wind | 7 | 720 | < th | dose independent | rect. | 14 | 9 |
| 636 | 264.40 | radiation | 10 | 1440 | > th | dose independent | rect. | 9 | 5 |
| 705 | 264.83 | radiation | 10 | 1440 | > th | dose independent | rect. | 10 | 3 |
| 775 | 265.17 | humidity | 90 | 720 | < th | dose dependent | rect. | 14 | 16 |
| 855 | 265.54 | humidity | 90 | 720 | < th | dose dependent | rect. | 15 | 10 |
| 894 | 265.72 | wind | 7 | 720 | < th | dose independent | rect. | 13 | 14 |
| 923 | 265.80 | humidity | 90 | 720 | < th | dose dependent | sin | 15 | NA |
| 954 | 265.94 | wind | 7 | 720 | < th | dose independent | sin | 16 | NA |
| 970 | 266.03 | humidity | 70 | 720 | > th | dose independent | rect. | 14 | 17 |
| 993 | 266.16 | humidity | 80 | 720 | > th | dose independent | sin | 20 | NA |