Os02g0210700
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Protein kinase-like domain containing protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0210700 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Protein kinase-like domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

Residual plot

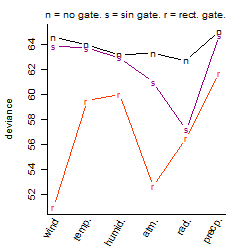
Process of the parameter reduction
(fixed parameters. wheather = radiation,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 61.94 | 0.37 | 3.03 | 0.0561 | 0.153 | 0.197 | -0.15 | 1.27 | 0.0466 | 6.43 | 40.1 | 44 | -- | -- |
| 65.03 | 0.376 | 3.03 | 0.0914 | 0.162 | 0.189 | -0.12 | -- | 0.0517 | 6.01 | 40.55 | 38 | -- | -- |
| 61.97 | 0.367 | 3.03 | 0.0544 | 0.153 | 0.203 | -- | 1.26 | 0.0471 | 6.25 | 40.46 | 43 | -- | -- |
| 65.06 | 0.376 | 3.03 | 0.0872 | 0.158 | 0.188 | -- | -- | 0.0537 | 6.26 | 40.49 | 38 | -- | -- |
| 63.18 | 0.37 | 3.04 | 0.058 | -- | 0.204 | -- | 1.28 | 0.0502 | -- | 40.41 | 43 | -- | -- |
| 65.75 | 0.38 | 3.03 | 0.0859 | 0.165 | -- | 0.0991 | -- | 0.052 | 7.72 | -- | -- | -- | -- |
| 65.77 | 0.38 | 3.03 | 0.088 | 0.167 | -- | -- | -- | 0.0516 | 7.47 | -- | -- | -- | -- |
| 66.35 | 0.379 | 3.03 | 0.0916 | -- | 0.191 | -- | -- | 0.057 | -- | 40.5 | 38 | -- | -- |
| 65.26 | 0.376 | 3.03 | -- | 0.159 | 0.189 | -- | -- | 0.044 | 6.23 | 40.42 | 38 | -- | -- |
| 67.25 | 0.383 | 3.04 | 0.0905 | -- | -- | -- | -- | 0.0543 | -- | -- | -- | -- | -- |
| 65.98 | 0.38 | 3.03 | -- | 0.167 | -- | -- | -- | 0.0418 | 7.43 | -- | -- | -- | -- |
| 66.57 | 0.38 | 3.04 | -- | -- | 0.191 | -- | -- | 0.0469 | -- | 40.51 | 38 | -- | -- |
| 67.47 | 0.383 | 3.04 | -- | -- | -- | -- | -- | 0.0442 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 50.95 | wind | 1 | 90 | < th | dose dependent | rect. | 8 | 2 |
| 3 | 51.37 | wind | 1 | 90 | < th | dose independent | rect. | 8 | 2 |
| 4 | 52.62 | atmosphere | 1010 | 90 | > th | dose independent | rect. | 8 | 1 |
| 5 | 52.62 | atmosphere | 1010 | 10 | > th | dose independent | rect. | 9 | 1 |
| 76 | 56.47 | radiation | 30 | 90 | > th | dose independent | rect. | 8 | 1 |
| 112 | 57.05 | radiation | 10 | 10 | > th | dose independent | rect. | 6 | 1 |
| 138 | 57.05 | radiation | 10 | 10 | > th | dose dependent | rect. | 6 | 1 |
| 164 | 57.19 | radiation | 40 | 30 | > th | dose independent | sin | 11 | NA |
| 166 | 57.26 | radiation | 40 | 30 | > th | dose independent | rect. | 14 | 20 |
| 228 | 57.69 | radiation | 20 | 90 | > th | dose dependent | rect. | 8 | 1 |
| 311 | 58.42 | wind | 1 | 270 | < th | dose independent | rect. | 6 | 1 |
| 376 | 58.57 | wind | 1 | 270 | < th | dose dependent | rect. | 6 | 1 |
| 395 | 58.66 | wind | 1 | 720 | < th | dose dependent | rect. | 4 | 1 |
| 400 | 58.76 | radiation | 40 | 30 | < th | dose independent | sin | 11 | NA |
| 441 | 58.95 | atmosphere | 1005 | 10 | > th | dose dependent | rect. | 9 | 1 |
| 463 | 59.01 | radiation | 30 | 10 | > th | dose dependent | rect. | 10 | 1 |
| 505 | 59.14 | radiation | 20 | 90 | > th | dose dependent | sin | 11 | NA |
| 523 | 59.48 | temperature | 25 | 10 | > th | dose independent | rect. | 8 | 3 |
| 575 | 59.60 | wind | 3 | 30 | < th | dose dependent | rect. | 6 | 1 |
| 616 | 59.82 | atmosphere | 1005 | 30 | > th | dose dependent | rect. | 6 | 1 |
| 743 | 60.02 | humidity | 80 | 30 | < th | dose independent | rect. | 9 | 1 |
| 814 | 60.22 | humidity | 80 | 30 | > th | dose dependent | rect. | 6 | 1 |
| 873 | 60.41 | temperature | 25 | 90 | < th | dose independent | rect. | 8 | 1 |
| 952 | 60.60 | temperature | 30 | 30 | < th | dose dependent | rect. | 6 | 1 |
| 956 | 60.62 | wind | 1 | 720 | < th | dose dependent | rect. | 4 | 3 |