Os02g0203300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : UDP-glucuronosyl/UDP-glucosyltransferase family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0203300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : UDP-glucuronosyl/UDP-glucosyltransferase family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
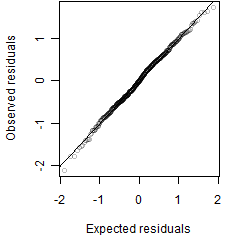
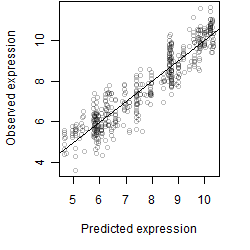 __
__

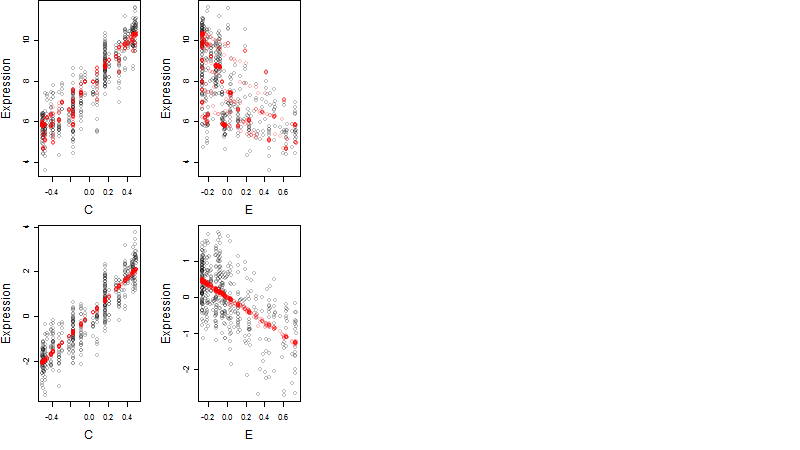
Dependence on each variable
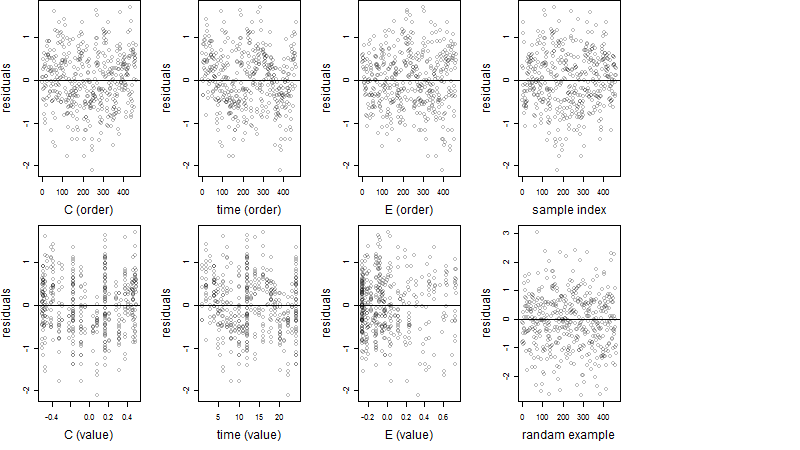
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose independent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 189.75 | 0.647 | 7.86 | -0.139 | 4.3 | -1.71 | 1.42 | 2.44 | -0.226 | 16.7 | 24.94 | 701 | 10.9 | -- |
| 197.41 | 0.654 | 7.81 | 0.329 | 4.2 | -1.67 | 0.0731 | -- | -0.091 | 16.7 | 24.94 | 625 | 9.95 | -- |
| 193.75 | 0.648 | 7.85 | 0.0112 | 4.31 | -1.63 | -- | 1.95 | -0.175 | 16.7 | 25 | 688 | 11 | -- |
| 197.42 | 0.654 | 7.81 | 0.332 | 4.21 | -1.67 | -- | -- | -0.0863 | 16.7 | 24.94 | 625 | 9.98 | -- |
| 261.17 | 0.753 | 7.65 | 0.628 | -- | -4.82 | -- | 0.266 | 0.33 | -- | 44.21 | 1099 | 10.7 | -- |
| 261.17 | 0.758 | 7.71 | 0.624 | 4.84 | -- | -0.272 | -- | 0.329 | 16.5 | -- | -- | -- | -- |
| 261.36 | 0.757 | 7.72 | 0.622 | 4.84 | -- | -- | -- | 0.327 | 16.5 | -- | -- | -- | -- |
| 261.36 | 0.753 | 7.65 | 0.622 | -- | -4.82 | -- | -- | 0.327 | -- | 48.53 | 1100 | 10.7 | -- |
| 200.22 | 0.659 | 7.82 | -- | 4.19 | -1.73 | -- | -- | -0.137 | 16.7 | 24.95 | 626 | 10 | -- |
| 1514.38 | 1.82 | 7.68 | 0.433 | -- | -- | -- | -- | 0.199 | -- | -- | -- | -- | -- |
| 271.56 | 0.771 | 7.73 | -- | 4.83 | -- | -- | -- | 0.258 | 16.5 | -- | -- | -- | -- |
| 271.56 | 0.768 | 7.67 | -- | -- | -4.81 | -- | -- | 0.258 | -- | 51.02 | 1107 | 10.7 | -- |
| 1519.33 | 1.82 | 7.69 | -- | -- | -- | -- | -- | 0.151 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance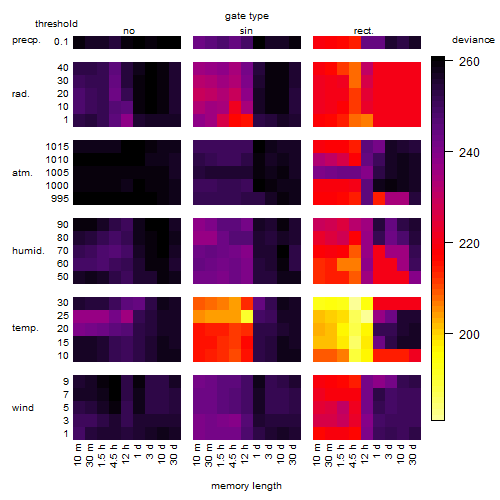
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 180.85 | temperature | 30 | 270 | < th | dose dependent | rect. | 16 | 6 |
| 2 | 181.94 | temperature | 25 | 720 | < th | dose independent | rect. | 14 | 10 |
| 14 | 185.13 | temperature | 15 | 270 | > th | dose dependent | rect. | 21 | 8 |
| 25 | 187.10 | temperature | 30 | 270 | < th | dose dependent | rect. | 16 | 16 |
| 50 | 190.35 | temperature | 25 | 720 | < th | dose independent | sin | 17 | NA |
| 61 | 191.31 | temperature | 25 | 720 | > th | dose independent | rect. | 14 | 10 |
| 75 | 192.53 | temperature | 30 | 10 | < th | dose dependent | rect. | 17 | 7 |
| 77 | 192.62 | temperature | 30 | 270 | < th | dose dependent | rect. | 19 | 1 |
| 164 | 196.77 | temperature | 10 | 720 | > th | dose dependent | rect. | 15 | 8 |
| 174 | 197.01 | temperature | 20 | 720 | > th | dose dependent | rect. | 15 | 8 |
| 238 | 199.07 | temperature | 25 | 720 | > th | dose independent | sin | 17 | NA |
| 378 | 202.51 | temperature | 20 | 270 | > th | dose independent | rect. | 21 | 7 |
| 445 | 203.82 | temperature | 25 | 90 | < th | dose dependent | sin | 13 | NA |
| 523 | 204.97 | temperature | 20 | 10 | > th | dose independent | rect. | 22 | 9 |
| 684 | 206.97 | radiation | 1 | 720 | < th | dose dependent | rect. | 16 | 8 |
| 685 | 206.98 | humidity | 60 | 90 | > th | dose independent | rect. | 19 | 16 |
| 689 | 207.03 | humidity | 60 | 270 | > th | dose independent | rect. | 17 | 16 |
| 712 | 207.30 | temperature | 10 | 270 | > th | dose dependent | rect. | 11 | 16 |
| 734 | 207.55 | radiation | 1 | 270 | > th | dose independent | rect. | 9 | 10 |
| 769 | 207.94 | radiation | 30 | 270 | < th | dose independent | rect. | 17 | 16 |
| 847 | 208.89 | radiation | 1 | 270 | < th | dose dependent | rect. | 16 | 5 |
| 855 | 209.02 | humidity | 50 | 270 | > th | dose dependent | rect. | 16 | 6 |
| 892 | 209.49 | temperature | 30 | 270 | < th | dose independent | rect. | 17 | 16 |
| 920 | 209.81 | radiation | 1 | 720 | < th | dose independent | rect. | 15 | 9 |
| 945 | 210.19 | temperature | 25 | 720 | < th | dose dependent | rect. | 21 | 1 |
| 952 | 210.27 | radiation | 1 | 270 | < th | dose independent | rect. | 15 | 6 |
| 990 | 210.72 | temperature | 10 | 270 | > th | dose dependent | sin | 14 | NA |
| 994 | 210.77 | temperature | 20 | 10 | > th | dose independent | rect. | 11 | 20 |