Os02g0202400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Plastidial ADP-glucose transporter.


FiT-DB / Search/ Help/ Sample detail
|
Os02g0202400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Plastidial ADP-glucose transporter.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

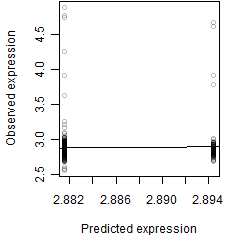 __
__
Dependence on each variable

Residual plot

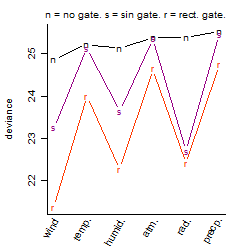
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24.02 | 0.23 | 2.87 | -0.0355 | 0.0592 | -0.197 | 0.0323 | 2.24 | 0.00914 | 8.47 | 0.09734 | 9 | -- | -- |
| 25.07 | 0.233 | 2.88 | -0.0532 | 0.0418 | 0.322 | -0.0306 | -- | 0.0193 | 8.54 | 0.7997 | 8 | -- | -- |
| 24.02 | 0.228 | 2.87 | -0.0346 | 0.0592 | -0.196 | -- | 2.23 | 0.00909 | 8.44 | 0.1103 | 8 | -- | -- |
| 25.06 | 0.233 | 2.88 | -0.0519 | 0.0442 | 0.33 | -- | -- | 0.0166 | 9.59 | 0.8 | 8 | -- | -- |
| 24.22 | 0.229 | 2.87 | -0.035 | -- | -0.178 | -- | 2.16 | 0.00983 | -- | 0.1043 | 8 | -- | -- |
| 25.72 | 0.238 | 2.88 | -0.033 | 0.0486 | -- | -0.0604 | -- | 0.00847 | 8.34 | -- | -- | -- | -- |
| 25.73 | 0.238 | 2.88 | -0.0353 | 0.0505 | -- | -- | -- | 0.00824 | 7.61 | -- | -- | -- | -- |
| 25.18 | 0.234 | 2.88 | -0.0501 | -- | 0.329 | -- | -- | 0.0172 | -- | 0.8 | 8 | -- | -- |
| 25.13 | 0.233 | 2.88 | -- | 0.0449 | 0.324 | -- | -- | 0.0226 | 9.62 | 0.7996 | 8 | -- | -- |
| 25.86 | 0.238 | 2.88 | -0.0346 | -- | -- | -- | -- | 0.00901 | -- | -- | -- | -- | -- |
| 25.76 | 0.237 | 2.88 | -- | 0.0502 | -- | -- | -- | 0.0122 | 7.66 | -- | -- | -- | -- |
| 25.24 | 0.234 | 2.88 | -- | -- | 0.319 | -- | -- | 0.0225 | -- | 0.8 | 8 | -- | -- |
| 25.89 | 0.238 | 2.88 | -- | -- | -- | -- | -- | 0.0128 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 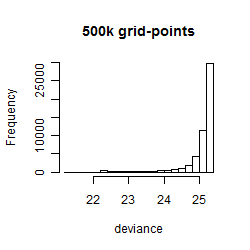
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 21.36 | wind | 9 | 10 | > th | dose dependent | rect. | 17 | 1 |
| 5 | 21.71 | wind | 1 | 10 | < th | dose independent | rect. | 6 | 4 |
| 13 | 21.85 | wind | 1 | 10 | < th | dose dependent | rect. | 7 | 17 |
| 19 | 21.91 | wind | 1 | 10 | < th | dose dependent | rect. | 7 | 3 |
| 43 | 22.10 | wind | 9 | 10 | > th | dose independent | rect. | 17 | 1 |
| 64 | 22.17 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 15 |
| 68 | 22.26 | humidity | 60 | 270 | < th | dose independent | rect. | 8 | 1 |
| 107 | 22.26 | humidity | 60 | 270 | < th | dose dependent | rect. | 8 | 1 |
| 146 | 22.28 | humidity | 60 | 10 | < th | dose independent | rect. | 9 | 1 |
| 216 | 22.28 | humidity | 60 | 90 | < th | dose independent | rect. | 18 | 15 |
| 224 | 22.28 | wind | 1 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 225 | 22.28 | humidity | 60 | 10 | < th | dose dependent | rect. | 9 | 1 |
| 283 | 22.28 | humidity | 60 | 90 | < th | dose dependent | rect. | 18 | 15 |
| 291 | 22.28 | humidity | 60 | 10 | < th | dose dependent | rect. | 17 | 17 |
| 353 | 22.31 | wind | 1 | 10 | < th | dose independent | rect. | 9 | 1 |
| 411 | 22.37 | humidity | 60 | 1440 | > th | dose independent | rect. | 22 | 11 |
| 437 | 22.39 | radiation | 40 | 30 | > th | dose dependent | rect. | 9 | 1 |
| 508 | 22.42 | humidity | 60 | 1440 | > th | dose independent | rect. | 24 | 9 |
| 533 | 22.49 | humidity | 60 | 1440 | > th | dose independent | rect. | 8 | 1 |
| 672 | 22.69 | radiation | 40 | 10 | > th | dose dependent | sin | 12 | NA |
| 712 | 22.78 | humidity | 60 | 1440 | > th | dose independent | rect. | 6 | 3 |
| 789 | 22.91 | radiation | 40 | 10 | > th | dose independent | rect. | 9 | 1 |