Os02g0187300
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, uncharacterized transcript.

FiT-DB / Search/ Help/ Sample detail
|
Os02g0187300 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, uncharacterized transcript.
|
|
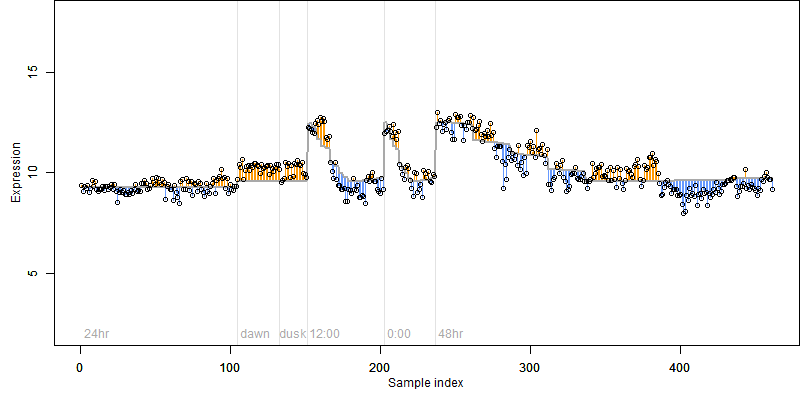
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
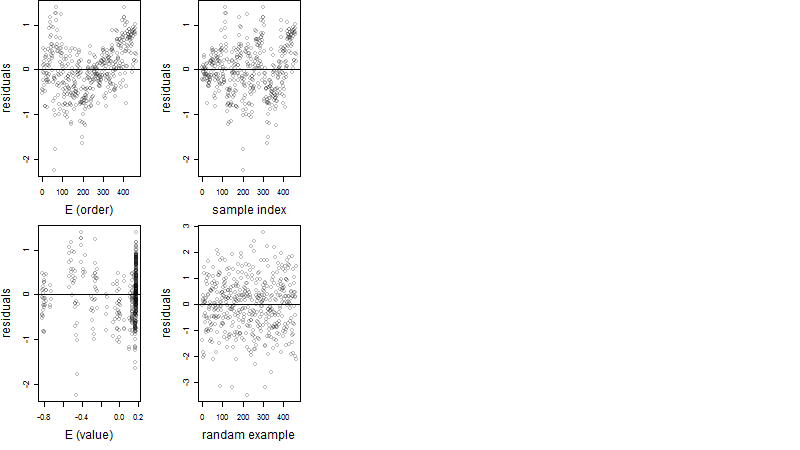
Dependence on each variable

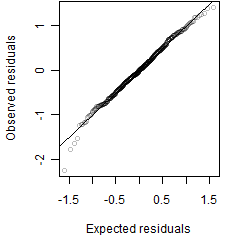
Residual plot
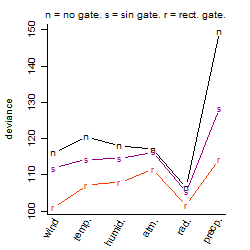
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 119.84 | 0.514 | 10 | -1.06 | 0.247 | -0.923 | 1.08 | 7.87 | -0.676 | 1.62 | 25.69 | 4716 | -- | -- |
| 123.76 | 0.518 | 10.2 | -1.36 | 0.242 | -1.99 | 1.27 | -- | -0.755 | 0.553 | 16.4 | 47520 | -- | -- |
| 122.53 | 0.516 | 10 | -0.928 | 0.241 | -0.828 | -- | 7.95 | -0.612 | 0.666 | 25.7 | 5508 | -- | -- |
| 127.66 | 0.526 | 10.2 | -1.33 | 0.247 | -2.09 | -- | -- | -0.722 | 0.944 | 16.47 | 47520 | -- | -- |
| 122.88 | 0.516 | 9.97 | -0.639 | -- | -0.714 | -- | 8.05 | -0.486 | -- | 25.5 | 6998 | -- | -- |
| 163.58 | 0.6 | 10.3 | -3.35 | 0.22 | -- | 1.09 | -- | -1.34 | 1.77 | -- | -- | -- | -- |
| 168.09 | 0.607 | 10.3 | -3.39 | 0.223 | -- | -- | -- | -1.34 | 0.892 | -- | -- | -- | -- |
| 131.45 | 0.534 | 10.2 | -1.34 | -- | -2.07 | -- | -- | -0.723 | -- | 16.47 | 47520 | -- | -- |
| 138.09 | 0.547 | 10.1 | -- | 0.265 | -2.96 | -- | -- | -0.294 | 0.928 | 18.1 | 47520 | -- | -- |
| 171.19 | 0.611 | 10.3 | -3.38 | -- | -- | -- | -- | -1.34 | -- | -- | -- | -- | -- |
| 470.56 | 1.01 | 10.3 | -- | 0.173 | -- | -- | -- | -0.967 | 0.0255 | -- | -- | -- | -- |
| 142.45 | 0.556 | 10.1 | -- | -- | -2.94 | -- | -- | -0.293 | -- | 18.02 | 47520 | -- | -- |
| 472.44 | 1.01 | 10.3 | -- | -- | -- | -- | -- | -0.965 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram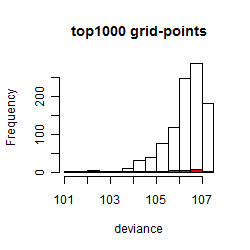 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 101.10 | wind | 9 | 43200 | < th | dose independent | rect. | 16 | 2 |
| 2 | 101.41 | wind | 9 | 43200 | > th | dose independent | rect. | 16 | 2 |
| 3 | 101.55 | radiation | 10 | 43200 | < th | dose independent | rect. | 8 | 1 |
| 4 | 102.05 | radiation | 10 | 43200 | < th | dose dependent | rect. | 7 | 1 |
| 6 | 102.37 | radiation | 10 | 43200 | < th | dose independent | rect. | 7 | 3 |
| 14 | 103.72 | wind | 1 | 14400 | < th | dose dependent | rect. | 22 | 5 |
| 17 | 103.84 | radiation | 1 | 43200 | > th | dose dependent | rect. | 7 | 1 |
| 28 | 104.21 | radiation | 20 | 43200 | < th | dose dependent | rect. | 22 | 12 |
| 71 | 104.94 | radiation | 20 | 43200 | > th | dose independent | rect. | 6 | 2 |
| 140 | 105.42 | radiation | 20 | 43200 | < th | dose dependent | sin | 9 | NA |
| 169 | 105.52 | radiation | 10 | 43200 | < th | dose independent | rect. | 21 | 14 |
| 184 | 105.65 | radiation | 20 | 43200 | < th | dose independent | rect. | 15 | 21 |
| 200 | 105.76 | radiation | 20 | 43200 | < th | dose independent | rect. | 17 | 19 |
| 220 | 105.84 | radiation | 20 | 43200 | > th | dose independent | rect. | 16 | 18 |
| 247 | 105.88 | radiation | 20 | 43200 | < th | dose independent | rect. | 7 | 9 |
| 329 | 106.06 | radiation | 20 | 43200 | > th | dose independent | sin | 11 | NA |
| 467 | 106.33 | radiation | 20 | 43200 | < th | dose independent | rect. | 17 | 23 |
| 481 | 106.35 | radiation | 20 | 43200 | < th | dose independent | sin | 1 | NA |
| 519 | 106.44 | radiation | 40 | 43200 | > th | dose independent | rect. | 13 | 1 |
| 556 | 106.52 | radiation | 20 | 43200 | < th | dose independent | no | NA | NA |
| 691 | 106.70 | radiation | 1 | 43200 | > th | dose dependent | sin | 9 | NA |
| 700 | 106.76 | radiation | 20 | 43200 | < th | dose independent | rect. | 14 | 1 |
| 710 | 106.80 | wind | 1 | 43200 | > th | dose independent | rect. | 22 | 4 |
| 782 | 106.94 | radiation | 30 | 43200 | < th | dose dependent | rect. | 5 | 11 |
| 785 | 106.95 | radiation | 40 | 43200 | > th | dose dependent | rect. | 13 | 1 |
| 818 | 107.00 | radiation | 10 | 43200 | > th | dose dependent | rect. | 15 | 19 |
| 832 | 107.06 | wind | 1 | 43200 | > th | dose independent | rect. | 20 | 6 |
| 851 | 107.10 | temperature | 20 | 14400 | > th | dose independent | rect. | 21 | 3 |