Os01g0832600
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Leucoanthocyanidin dioxygenase-like protein.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0832600 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Leucoanthocyanidin dioxygenase-like protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

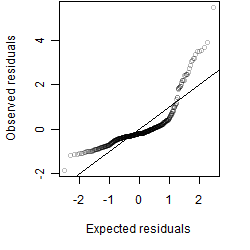
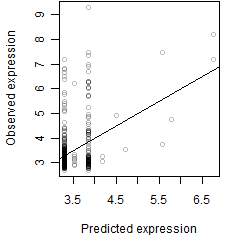
Dependence on each variable

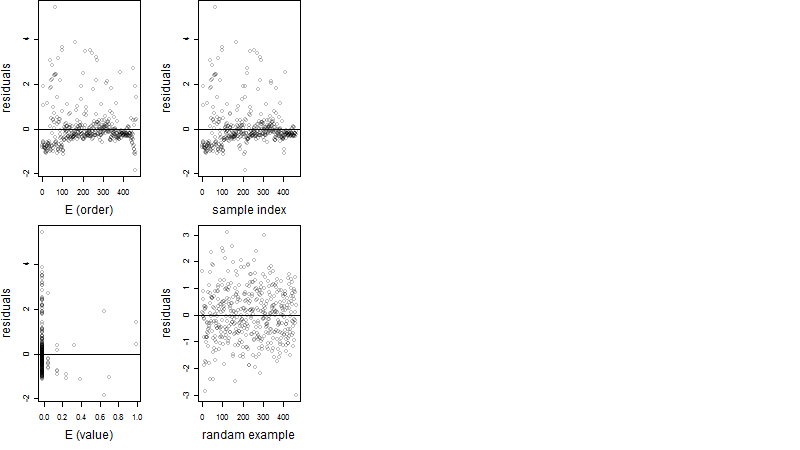
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 337.12 | 0.863 | 3.34 | 0.00335 | 0.367 | 1.6 | -0.673 | -7.92 | 0.551 | 21 | 10.31 | 569 | -- | -- |
| 339.11 | 0.858 | 3.36 | 0.08 | 0.358 | 3.21 | -0.708 | -- | 0.56 | 21.2 | 10.35 | 585 | -- | -- |
| 338.53 | 0.857 | 3.33 | 0.0199 | 0.366 | 1.52 | -- | -8.7 | 0.554 | 21.6 | 10.31 | 550 | -- | -- |
| 340.72 | 0.86 | 3.36 | 0.104 | 0.355 | 3.32 | -- | -- | 0.547 | 21.4 | 10.39 | 585 | -- | -- |
| 346.25 | 0.867 | 3.33 | 0.034 | -- | 1.78 | -- | -8.34 | 0.555 | -- | 10.35 | 557 | -- | -- |
| 376.48 | 0.91 | 3.38 | -0.113 | 0.472 | -- | -1.25 | -- | 0.469 | 20.3 | -- | -- | -- | -- |
| 380.98 | 0.914 | 3.38 | -0.0792 | 0.474 | -- | -- | -- | 0.467 | 21 | -- | -- | -- | -- |
| 347.98 | 0.869 | 3.35 | 0.116 | -- | 3.5 | -- | -- | 0.55 | -- | 10.4 | 580 | -- | -- |
| 341 | 0.86 | 3.37 | -- | 0.357 | 3.28 | -- | -- | 0.534 | 21.4 | 10.4 | 577 | -- | -- |
| 393.88 | 0.927 | 3.37 | -0.0786 | -- | -- | -- | -- | 0.464 | -- | -- | -- | -- | -- |
| 381.15 | 0.913 | 3.38 | -- | 0.474 | -- | -- | -- | 0.476 | 21 | -- | -- | -- | -- |
| 348.33 | 0.869 | 3.35 | -- | -- | 3.46 | -- | -- | 0.536 | -- | 10.4 | 584 | -- | -- |
| 394.04 | 0.927 | 3.37 | -- | -- | -- | -- | -- | 0.473 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance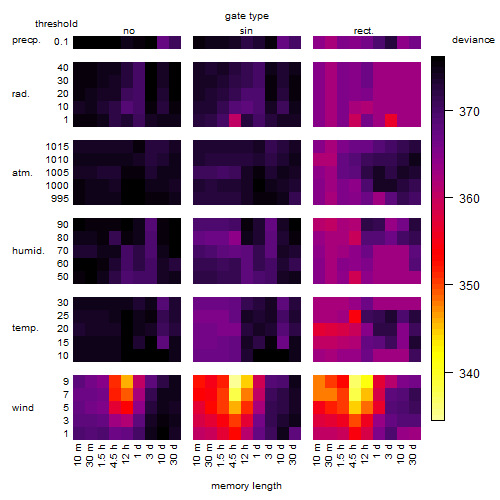
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 334.42 | wind | 9 | 270 | > th | dose independent | sin | 13 | NA |
| 4 | 335.90 | wind | 9 | 270 | > th | dose dependent | sin | 13 | NA |
| 7 | 336.91 | wind | 9 | 270 | > th | dose independent | rect. | 15 | 11 |
| 15 | 337.01 | wind | 9 | 270 | > th | dose independent | rect. | 15 | 8 |
| 32 | 338.01 | wind | 7 | 270 | > th | dose dependent | rect. | 14 | 12 |
| 42 | 338.14 | wind | 9 | 270 | > th | dose dependent | rect. | 15 | 8 |
| 128 | 339.43 | wind | 9 | 270 | < th | dose independent | sin | 14 | NA |
| 160 | 341.16 | wind | 9 | 720 | > th | dose dependent | rect. | 15 | 1 |
| 161 | 341.37 | wind | 9 | 720 | > th | dose dependent | rect. | 12 | 5 |
| 232 | 342.73 | wind | 9 | 720 | > th | dose independent | rect. | 12 | 4 |
| 240 | 342.89 | wind | 9 | 270 | > th | dose independent | rect. | 15 | 3 |
| 268 | 343.31 | wind | 9 | 720 | < th | dose independent | rect. | 3 | 16 |
| 274 | 343.67 | wind | 9 | 720 | > th | dose dependent | rect. | 22 | 19 |
| 277 | 343.77 | wind | 9 | 720 | > th | dose dependent | rect. | 9 | 8 |
| 291 | 343.86 | wind | 9 | 720 | < th | dose independent | rect. | 17 | 23 |
| 337 | 344.78 | wind | 9 | 720 | > th | dose dependent | sin | 22 | NA |
| 345 | 344.99 | wind | 9 | 720 | < th | dose independent | sin | 23 | NA |
| 396 | 345.17 | wind | 9 | 720 | > th | dose independent | rect. | 22 | 19 |
| 442 | 345.36 | wind | 9 | 720 | > th | dose independent | rect. | 9 | 8 |
| 465 | 345.54 | wind | 9 | 720 | < th | dose independent | sin | 20 | NA |
| 507 | 345.68 | wind | 9 | 720 | > th | dose dependent | no | NA | NA |
| 592 | 346.01 | wind | 9 | 720 | > th | dose independent | sin | 23 | NA |
| 712 | 346.55 | wind | 9 | 720 | < th | dose independent | no | NA | NA |
| 806 | 346.89 | wind | 9 | 720 | > th | dose independent | no | NA | NA |
| 953 | 347.69 | wind | 9 | 10 | > th | dose independent | rect. | 17 | 3 |
| 988 | 347.81 | wind | 7 | 30 | > th | dose dependent | rect. | 16 | 5 |