Os01g0760800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to Homeodomain protein 1.

FiT-DB / Search/ Help/ Sample detail
|
Os01g0760800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Homeodomain protein 1.
|
|
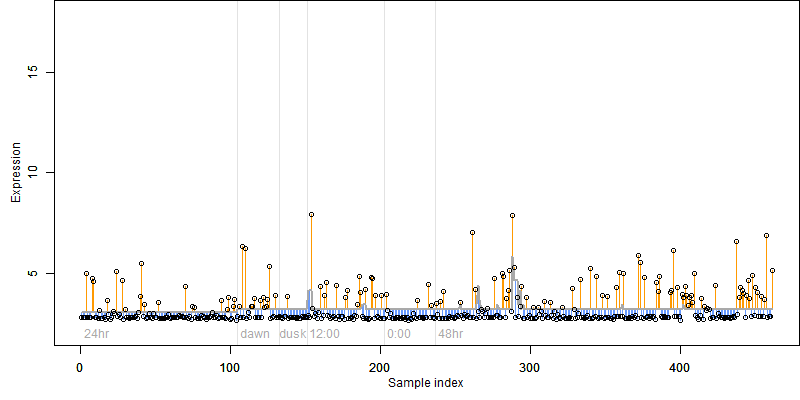
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

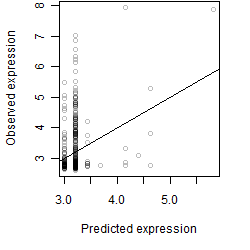 __
__
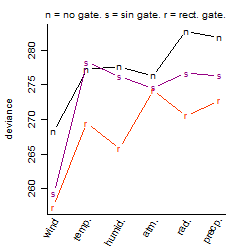
Dependence on each variable

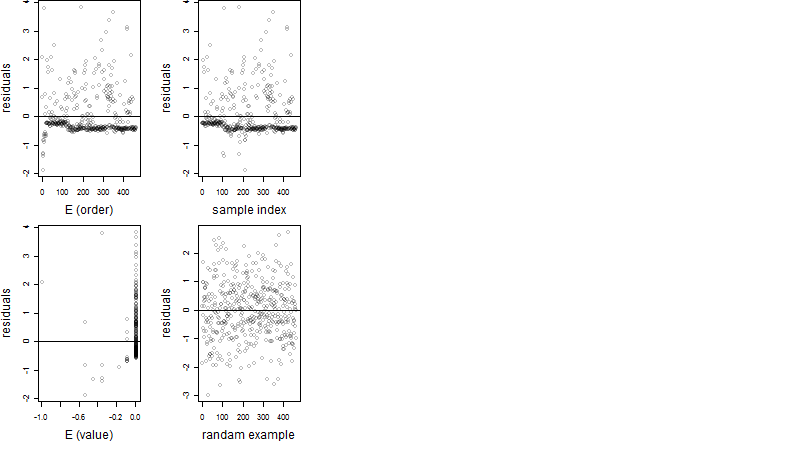
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = wind,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 266.82 | 0.767 | 3.2 | 0.362 | 0.385 | -2.24 | 0.0242 | 2.65 | -0.146 | 7.71 | 8.977 | 31 | -- | -- |
| 267.09 | 0.761 | 3.21 | 0.364 | 0.39 | -2.94 | 0.078 | -- | -0.149 | 7.51 | 8.9 | 31 | -- | -- |
| 266.82 | 0.761 | 3.2 | 0.362 | 0.385 | -2.24 | -- | 2.66 | -0.146 | 7.71 | 8.977 | 31 | -- | -- |
| 267.06 | 0.761 | 3.21 | 0.377 | 0.385 | -2.93 | -- | -- | -0.147 | 7.72 | 8.929 | 31 | -- | -- |
| 274.8 | 0.772 | 3.21 | 0.362 | -- | -2.14 | -- | 2.76 | -0.142 | -- | 8.913 | 31 | -- | -- |
| 288.43 | 0.796 | 3.23 | 0.211 | 0.36 | -- | -0.117 | -- | -0.21 | 8.81 | -- | -- | -- | -- |
| 288.46 | 0.795 | 3.23 | 0.207 | 0.362 | -- | -- | -- | -0.211 | 8.66 | -- | -- | -- | -- |
| 275.06 | 0.772 | 3.22 | 0.378 | -- | -2.85 | -- | -- | -0.144 | -- | 8.985 | 31 | -- | -- |
| 270.68 | 0.766 | 3.22 | -- | 0.386 | -2.7 | -- | -- | -0.193 | 7.72 | 8.923 | 31 | -- | -- |
| 295.84 | 0.804 | 3.24 | 0.208 | -- | -- | -- | -- | -0.208 | -- | -- | -- | -- | -- |
| 289.59 | 0.796 | 3.23 | -- | 0.362 | -- | -- | -- | -0.234 | 8.61 | -- | -- | -- | -- |
| 278.69 | 0.778 | 3.23 | -- | -- | -2.62 | -- | -- | -0.189 | -- | 8.935 | 31 | -- | -- |
| 296.98 | 0.804 | 3.24 | -- | -- | -- | -- | -- | -0.231 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 257.31 | wind | 9 | 30 | > th | dose dependent | rect. | 11 | 3 |
| 29 | 258.14 | wind | 9 | 30 | > th | dose dependent | rect. | 18 | 20 |
| 37 | 258.45 | wind | 9 | 30 | > th | dose independent | rect. | 11 | 3 |
| 58 | 259.30 | wind | 9 | 30 | > th | dose dependent | sin | 6 | NA |
| 107 | 260.45 | wind | 9 | 30 | < th | dose independent | sin | 5 | NA |
| 118 | 260.96 | wind | 9 | 30 | > th | dose independent | sin | 5 | NA |
| 307 | 265.76 | humidity | 60 | 4320 | < th | dose independent | rect. | 8 | 1 |
| 340 | 266.09 | humidity | 60 | 4320 | < th | dose dependent | rect. | 8 | 1 |
| 494 | 268.27 | wind | 9 | 30 | < th | dose independent | no | NA | NA |
| 520 | 268.49 | humidity | 60 | 4320 | > th | dose independent | rect. | 21 | 12 |
| 553 | 268.73 | humidity | 60 | 4320 | > th | dose independent | rect. | 24 | 9 |
| 671 | 269.28 | wind | 9 | 30 | > th | dose independent | no | NA | NA |
| 712 | 269.28 | wind | 9 | 30 | > th | dose independent | rect. | 13 | 23 |
| 719 | 269.29 | humidity | 60 | 4320 | > th | dose independent | rect. | 3 | 6 |
| 727 | 269.47 | temperature | 25 | 10 | < th | dose dependent | rect. | 24 | 2 |
| 736 | 269.59 | humidity | 60 | 4320 | > th | dose independent | rect. | 8 | 1 |
| 749 | 269.64 | temperature | 20 | 10 | < th | dose independent | rect. | 2 | 1 |
| 807 | 270.51 | radiation | 10 | 270 | < th | dose dependent | rect. | 10 | 1 |