Os01g0757200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Similar to GA 2-oxidase 4.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0757200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to GA 2-oxidase 4.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
Dependence on each variable

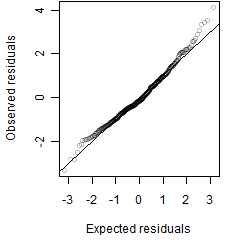
Residual plot

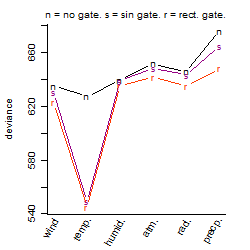
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = < th, dose dependency = dose dependent, type of G = sin)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 548.04 | 1.1 | 5.2 | 0.807 | 4.71 | 4.12 | -0.765 | -3.2 | 0.833 | 18 | 29.71 | 256 | 22.8 | -- |
| 560.76 | 1.1 | 5.21 | 0.7 | 4.27 | 4.28 | -0.0883 | -- | 0.753 | 17.8 | 27.69 | 294 | 22.6 | -- |
| 546.2 | 1.09 | 5.18 | 0.775 | 4.33 | 4.18 | -- | -2.63 | 0.812 | 17.9 | 28.72 | 220 | 21.9 | -- |
| 555.64 | 1.1 | 5.22 | 0.551 | 4.23 | 4.51 | -- | -- | 0.75 | 17.9 | 28.05 | 220 | 21.6 | -- |
| 710.67 | 1.24 | 5.1 | 1.03 | -- | 4.93 | -- | -4.42 | 0.737 | -- | 67.6 | 1134 | 22.7 | -- |
| 714.62 | 1.25 | 5.3 | -0.167 | 3.23 | -- | -0.255 | -- | 0.224 | 16.8 | -- | -- | -- | -- |
| 714.78 | 1.25 | 5.3 | -0.169 | 3.23 | -- | -- | -- | 0.223 | 16.8 | -- | -- | -- | -- |
| 682.64 | 1.22 | 5.2 | 0.224 | -- | 4.32 | -- | -- | 0.484 | -- | 125.1 | 1236 | 23.4 | -- |
| 562.3 | 1.1 | 5.24 | -- | 4.2 | 4.25 | -- | -- | 0.661 | 17.8 | 28.2 | 210 | 21.6 | -- |
| 1267.97 | 1.66 | 5.27 | -0.29 | -- | -- | -- | -- | 0.139 | -- | -- | -- | -- | -- |
| 715.53 | 1.25 | 5.3 | -- | 3.24 | -- | -- | -- | 0.242 | 16.8 | -- | -- | -- | -- |
| 683.64 | 1.22 | 5.21 | -- | -- | 4.24 | -- | -- | 0.441 | -- | 148.5 | 1257 | 23.6 | -- |
| 1270.19 | 1.66 | 5.26 | -- | -- | -- | -- | -- | 0.171 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance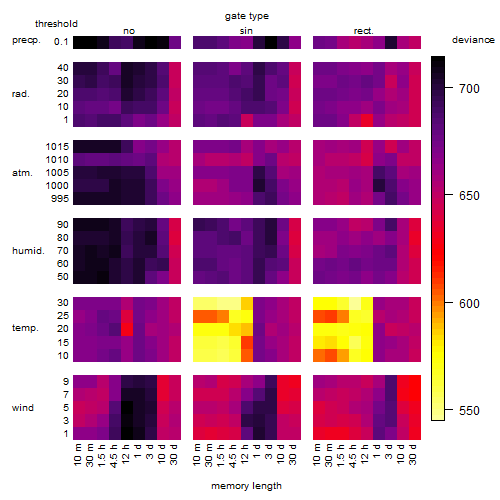
|
Histogram 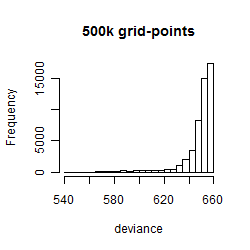
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 544.95 | temperature | 30 | 270 | < th | dose dependent | rect. | 23 | 16 |
| 3 | 548.93 | temperature | 30 | 270 | < th | dose dependent | sin | 5 | NA |
| 5 | 549.81 | temperature | 30 | 90 | < th | dose dependent | sin | 3 | NA |
| 22 | 557.91 | temperature | 10 | 90 | > th | dose dependent | sin | 3 | NA |
| 24 | 558.11 | temperature | 15 | 270 | > th | dose dependent | rect. | 2 | 12 |
| 50 | 563.86 | temperature | 25 | 720 | < th | dose dependent | rect. | 1 | 9 |
| 79 | 566.94 | temperature | 25 | 720 | < th | dose dependent | rect. | 3 | 5 |
| 95 | 568.57 | temperature | 25 | 270 | < th | dose dependent | rect. | 5 | 10 |
| 120 | 570.34 | temperature | 25 | 720 | < th | dose dependent | rect. | 5 | 3 |
| 137 | 571.57 | temperature | 20 | 10 | > th | dose dependent | rect. | 3 | 13 |
| 152 | 572.47 | temperature | 15 | 720 | > th | dose dependent | rect. | 2 | 4 |
| 196 | 574.55 | temperature | 30 | 10 | < th | dose dependent | rect. | 7 | 11 |
| 197 | 574.56 | temperature | 30 | 90 | < th | dose dependent | rect. | 7 | 10 |
| 210 | 575.13 | temperature | 10 | 720 | > th | dose dependent | rect. | 23 | 7 |
| 214 | 575.49 | temperature | 20 | 720 | < th | dose dependent | rect. | 3 | 2 |
| 274 | 579.28 | temperature | 20 | 720 | < th | dose independent | rect. | 2 | 8 |
| 298 | 580.42 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 8 |
| 313 | 581.04 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 5 |
| 341 | 581.58 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 11 |
| 347 | 581.78 | temperature | 20 | 720 | > th | dose independent | rect. | 3 | 3 |
| 381 | 583.13 | temperature | 30 | 10 | < th | dose dependent | rect. | 7 | 9 |
| 587 | 588.83 | temperature | 20 | 720 | < th | dose independent | rect. | 3 | 2 |
| 706 | 592.04 | temperature | 30 | 10 | < th | dose dependent | rect. | 7 | 7 |
| 730 | 592.49 | temperature | 20 | 720 | < th | dose independent | sin | 5 | NA |
| 833 | 595.47 | temperature | 30 | 30 | < th | dose dependent | rect. | 9 | 9 |
| 910 | 597.31 | temperature | 20 | 10 | > th | dose dependent | rect. | 24 | 16 |
| 993 | 599.57 | temperature | 20 | 720 | > th | dose independent | sin | 5 | NA |