Os01g0659400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, putative npRNA.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0659400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, putative npRNA.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
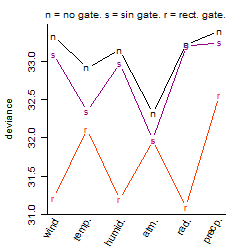
Dependence on each variable

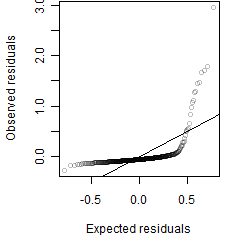
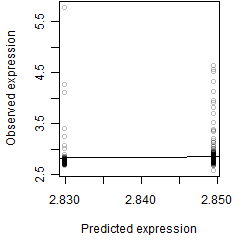
Residual plot

Process of the parameter reduction
(fixed parameters. wheather = atmosphere,
response mode = < th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 32.32 | 0.265 | 26.7 | -146 | 0.0395 | 6990 | -0.0374 | -42700 | -0.0172 | 4.97 | 995 | 10 | -- | -- |
| 31.98 | 0.265 | 2.85 | -0.00966 | 0.0369 | 1.31 | -0.0446 | -- | -0.0179 | 4.79 | 994.8 | 20 | -- | -- |
| 32.32 | 0.265 | 26.7 | -146 | 0.0394 | 6990 | -- | -42700 | -0.0174 | 4.88 | 995 | 10 | -- | -- |
| 31.98 | 0.263 | 2.85 | -0.00999 | 0.0374 | 1.31 | -- | -- | -0.0176 | 4.69 | 994.8 | 20 | -- | -- |
| 32.4 | 0.265 | 26.7 | -146 | -- | 6990 | -- | -42700 | -0.0161 | -- | 995 | 10 | -- | -- |
| 33.67 | 0.272 | 2.85 | -0.00215 | 0.0247 | -- | -0.0873 | -- | -0.0207 | 5.11 | -- | -- | -- | -- |
| 33.69 | 0.272 | 2.85 | -0.00214 | 0.0241 | -- | -- | -- | -0.0203 | 4.75 | -- | -- | -- | -- |
| 32.05 | 0.264 | 2.85 | -0.00848 | -- | 1.29 | -- | -- | -0.0166 | -- | 994.8 | 20 | -- | -- |
| 31.98 | 0.263 | 2.85 | -- | 0.0371 | 1.31 | -- | -- | -0.0165 | 4.7 | 994.8 | 20 | -- | -- |
| 33.72 | 0.271 | 2.85 | -0.00123 | -- | -- | -- | -- | -0.0197 | -- | -- | -- | -- | -- |
| 33.69 | 0.271 | 2.85 | -- | 0.0241 | -- | -- | -- | -0.0201 | 4.75 | -- | -- | -- | -- |
| 32.05 | 0.264 | 2.85 | -- | -- | 1.29 | -- | -- | -0.0157 | -- | 994.8 | 20 | -- | -- |
| 33.72 | 0.271 | 2.85 | -- | -- | -- | -- | -- | -0.0195 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance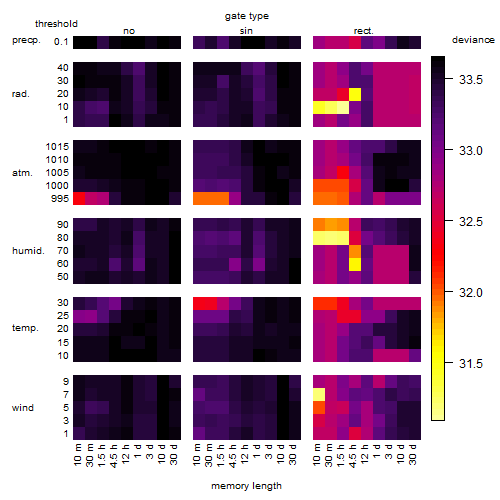
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 31.09 | radiation | 10 | 90 | < th | dose dependent | rect. | 8 | 2 |
| 2 | 31.19 | humidity | 80 | 30 | < th | dose dependent | rect. | 19 | 1 |
| 7 | 31.21 | wind | 7 | 10 | > th | dose dependent | rect. | 9 | 1 |
| 63 | 31.51 | radiation | 20 | 270 | > th | dose dependent | rect. | 16 | 1 |
| 89 | 31.60 | humidity | 60 | 270 | < th | dose independent | rect. | 16 | 1 |
| 112 | 31.75 | wind | 7 | 10 | > th | dose independent | rect. | 9 | 1 |
| 122 | 31.77 | radiation | 10 | 90 | < th | dose independent | rect. | 8 | 2 |
| 123 | 31.77 | humidity | 60 | 270 | < th | dose dependent | rect. | 16 | 1 |
| 156 | 31.97 | atmosphere | 995 | 10 | > th | dose independent | sin | 6 | NA |
| 159 | 31.97 | atmosphere | 995 | 10 | < th | dose dependent | sin | 5 | NA |
| 162 | 31.97 | atmosphere | 995 | 10 | < th | dose dependent | rect. | 18 | 23 |
| 166 | 31.98 | atmosphere | 995 | 10 | < th | dose independent | sin | 5 | NA |
| 168 | 31.98 | atmosphere | 995 | 90 | < th | dose independent | sin | 6 | NA |
| 169 | 31.98 | atmosphere | 995 | 90 | < th | dose independent | rect. | 14 | 1 |
| 170 | 31.98 | atmosphere | 995 | 10 | < th | dose independent | rect. | 15 | 1 |
| 299 | 31.98 | atmosphere | 995 | 90 | < th | dose dependent | rect. | 14 | 1 |
| 300 | 31.98 | atmosphere | 995 | 10 | < th | dose dependent | rect. | 15 | 1 |
| 520 | 32.11 | temperature | 30 | 30 | > th | dose dependent | rect. | 16 | 1 |
| 557 | 32.15 | humidity | 80 | 30 | < th | dose independent | rect. | 19 | 1 |
| 569 | 32.16 | temperature | 30 | 10 | > th | dose dependent | rect. | 15 | 3 |
| 658 | 32.22 | atmosphere | 995 | 10 | > th | dose independent | rect. | 3 | 23 |
| 659 | 32.22 | atmosphere | 995 | 30 | > th | dose independent | rect. | 15 | 23 |
| 716 | 32.32 | atmosphere | 1005 | 90 | < th | dose independent | rect. | 18 | 1 |
| 717 | 32.32 | atmosphere | 995 | 10 | < th | dose dependent | no | NA | NA |
| 951 | 32.35 | temperature | 30 | 10 | > th | dose dependent | sin | 12 | NA |
| 961 | 32.36 | atmosphere | 995 | 10 | < th | dose dependent | sin | 18 | NA |
| 981 | 32.43 | temperature | 25 | 90 | > th | dose independent | rect. | 1 | 3 |
| 987 | 32.44 | temperature | 25 | 270 | > th | dose independent | rect. | 1 | 1 |
| 994 | 32.46 | temperature | 25 | 90 | > th | dose dependent | rect. | 2 | 2 |