Os01g0651200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Lipase, class 3 family protein.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0651200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lipase, class 3 family protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
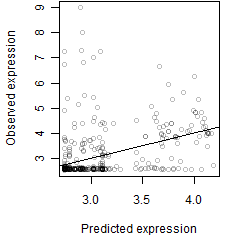 __
__
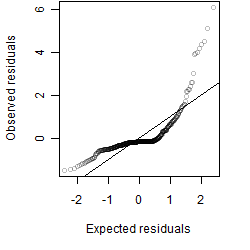
Dependence on each variable

Residual plot

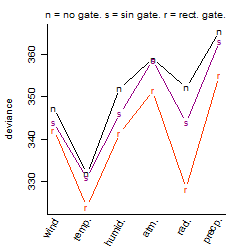
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 324.65 | 0.847 | 3.15 | -0.955 | 0.0827 | 1.86 | -0.699 | -4.22 | -0.215 | 10.4 | 26.9 | 1022 | -- | -- |
| 326.85 | 0.842 | 3.05 | -0.178 | 0.137 | 1.58 | -0.346 | -- | -0.0967 | 10.4 | 26.34 | 959 | -- | -- |
| 326.15 | 0.841 | 3.12 | -0.793 | 0.118 | 1.76 | -- | -3.1 | -0.181 | 13.4 | 26.89 | 1031 | -- | -- |
| 327.48 | 0.843 | 3.06 | -0.202 | 0.121 | 1.55 | -- | -- | -0.0966 | 11.8 | 26.33 | 968 | -- | -- |
| 326.88 | 0.842 | 3.13 | -0.829 | -- | 1.76 | -- | -3.48 | -0.176 | -- | 27.07 | 1024 | -- | -- |
| 390.74 | 0.927 | 3.06 | 0.214 | 0.279 | -- | 0.87 | -- | -0.0681 | 21.7 | -- | -- | -- | -- |
| 393.55 | 0.929 | 3.06 | 0.182 | 0.279 | -- | -- | -- | -0.0689 | 21.3 | -- | -- | -- | -- |
| 328.17 | 0.844 | 3.06 | -0.191 | -- | 1.48 | -- | -- | -0.0882 | -- | 26.59 | 972 | -- | -- |
| 328.43 | 0.844 | 3.05 | -- | 0.102 | 1.49 | -- | -- | -0.0697 | 11.7 | 26.5 | 968 | -- | -- |
| 398.08 | 0.932 | 3.05 | 0.183 | -- | -- | -- | -- | -0.07 | -- | -- | -- | -- | -- |
| 394.42 | 0.929 | 3.06 | -- | 0.279 | -- | -- | -- | -0.0892 | 21.4 | -- | -- | -- | -- |
| 328.99 | 0.845 | 3.05 | -- | -- | 1.44 | -- | -- | -0.0556 | -- | 26.76 | 969 | -- | -- |
| 398.97 | 0.932 | 3.06 | -- | -- | -- | -- | -- | -0.0904 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram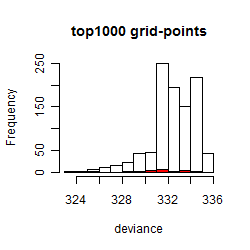 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 323.97 | temperature | 10 | 270 | > th | dose dependent | rect. | 13 | 14 |
| 3 | 324.97 | temperature | 10 | 270 | > th | dose dependent | rect. | 22 | 21 |
| 4 | 325.10 | temperature | 25 | 720 | > th | dose dependent | rect. | 15 | 20 |
| 38 | 328.19 | radiation | 30 | 1440 | < th | dose independent | rect. | 19 | 13 |
| 49 | 328.80 | temperature | 10 | 720 | > th | dose dependent | rect. | 7 | 20 |
| 75 | 329.80 | radiation | 30 | 1440 | > th | dose independent | rect. | 7 | 1 |
| 105 | 330.17 | radiation | 30 | 1440 | < th | dose independent | rect. | 7 | 1 |
| 113 | 330.56 | temperature | 25 | 1440 | > th | dose dependent | rect. | 16 | 1 |
| 114 | 330.56 | temperature | 25 | 1440 | > th | dose dependent | rect. | 11 | 7 |
| 141 | 330.91 | temperature | 25 | 1440 | > th | dose dependent | rect. | 11 | 12 |
| 151 | 331.06 | temperature | 25 | 1440 | > th | dose dependent | sin | 20 | NA |
| 194 | 331.31 | temperature | 25 | 720 | > th | dose dependent | sin | 17 | NA |
| 204 | 331.35 | temperature | 25 | 1440 | > th | dose dependent | rect. | 21 | 21 |
| 329 | 331.74 | radiation | 1 | 720 | > th | dose dependent | rect. | 17 | 13 |
| 371 | 331.88 | temperature | 25 | 1440 | > th | dose dependent | no | NA | NA |
| 394 | 331.98 | temperature | 25 | 720 | > th | dose dependent | sin | 10 | NA |
| 557 | 332.79 | temperature | 30 | 720 | > th | dose independent | sin | 11 | NA |
| 606 | 333.13 | temperature | 30 | 720 | > th | dose independent | rect. | 17 | 17 |
| 672 | 333.60 | temperature | 30 | 720 | < th | dose independent | sin | 11 | NA |
| 717 | 333.85 | temperature | 10 | 720 | > th | dose dependent | sin | 19 | NA |
| 762 | 334.11 | temperature | 30 | 720 | > th | dose independent | rect. | 15 | 19 |
| 886 | 334.75 | temperature | 15 | 720 | > th | dose dependent | rect. | 15 | 4 |