Os01g0633800
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Non-protein coding transcript, unclassifiable transcript.


FiT-DB / Search/ Help/ Sample detail
|
Os01g0633800 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Non-protein coding transcript, unclassifiable transcript.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
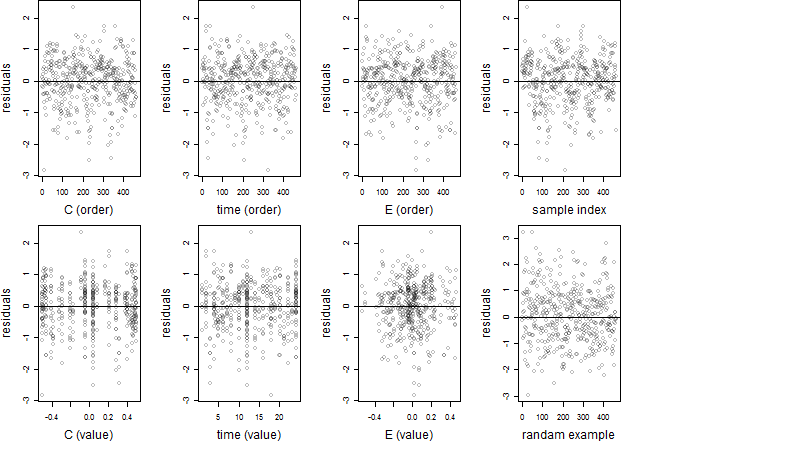
Dependence on each variable

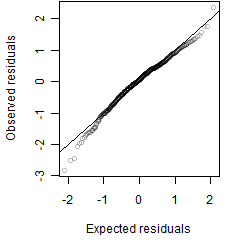
Residual plot
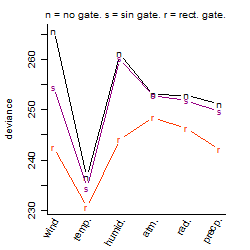
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 234.97 | 0.72 | 5.99 | 0.294 | 0.722 | 1.64 | -0.223 | 3.42 | -0.572 | 7.7 | 9.233 | 1715 | -- | -- |
| 246.3 | 0.731 | 6.06 | -0.285 | 1.2 | 1.97 | 0.107 | -- | -0.643 | 7.7 | 13.62 | 605 | -- | -- |
| 235.08 | 0.714 | 5.98 | 0.29 | 0.731 | 1.64 | -- | 3.45 | -0.57 | 7.75 | 8.67 | 1740 | -- | -- |
| 243.66 | 0.727 | 6.07 | -0.385 | 1.17 | 2.13 | -- | -- | -0.765 | 6.27 | 10.06 | 264 | -- | -- |
| 259.84 | 0.751 | 5.97 | 0.368 | -- | 1.5 | -- | 3.64 | -0.512 | -- | 5.715 | 2962 | -- | -- |
| 290.89 | 0.8 | 6.02 | 0.0762 | 0.664 | -- | -0.334 | -- | -0.464 | 7.6 | -- | -- | -- | -- |
| 291.17 | 0.799 | 6.02 | 0.0691 | 0.662 | -- | -- | -- | -0.463 | 7.75 | -- | -- | -- | -- |
| 267.17 | 0.761 | 6.1 | -0.458 | -- | 1.66 | -- | -- | -0.758 | -- | 14.6 | 1444 | -- | -- |
| 247.2 | 0.732 | 6.06 | -- | 1.12 | 1.94 | -- | -- | -0.7 | 6.37 | 9.403 | 267 | -- | -- |
| 314.85 | 0.829 | 6.03 | 0.0774 | -- | -- | -- | -- | -0.454 | -- | -- | -- | -- | -- |
| 291.3 | 0.798 | 6.02 | -- | 0.663 | -- | -- | -- | -0.471 | 7.74 | -- | -- | -- | -- |
| 271.89 | 0.768 | 6.08 | -- | -- | 1.47 | -- | -- | -0.683 | -- | 8.074 | 1443 | -- | -- |
| 315.01 | 0.828 | 6.03 | -- | -- | -- | -- | -- | -0.462 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 230.64 | temperature | 20 | 4320 | > th | dose dependent | rect. | 23 | 1 |
| 3 | 231.44 | temperature | 20 | 4320 | > th | dose dependent | rect. | 21 | 4 |
| 12 | 231.80 | temperature | 10 | 30 | > th | dose dependent | rect. | 3 | 23 |
| 43 | 232.94 | temperature | 25 | 4320 | < th | dose dependent | rect. | 21 | 1 |
| 49 | 233.11 | temperature | 20 | 4320 | > th | dose dependent | rect. | 3 | 1 |
| 73 | 233.57 | temperature | 30 | 4320 | < th | dose dependent | rect. | 21 | 3 |
| 78 | 233.64 | temperature | 30 | 4320 | < th | dose dependent | rect. | 20 | 6 |
| 93 | 233.85 | temperature | 25 | 4320 | < th | dose independent | rect. | 21 | 12 |
| 95 | 233.88 | temperature | 30 | 4320 | < th | dose dependent | rect. | 23 | 1 |
| 122 | 234.18 | temperature | 25 | 4320 | > th | dose independent | rect. | 21 | 12 |
| 148 | 234.44 | temperature | 20 | 4320 | > th | dose dependent | sin | 11 | NA |
| 219 | 234.97 | temperature | 15 | 10 | > th | dose dependent | rect. | 21 | 23 |
| 279 | 235.38 | temperature | 15 | 270 | > th | dose dependent | rect. | 3 | 23 |
| 301 | 235.51 | temperature | 30 | 270 | < th | dose dependent | rect. | 23 | 23 |
| 340 | 235.67 | temperature | 30 | 4320 | < th | dose dependent | sin | 11 | NA |
| 353 | 235.75 | temperature | 10 | 1440 | > th | dose dependent | rect. | 20 | 19 |
| 363 | 235.78 | temperature | 10 | 1440 | > th | dose dependent | rect. | 22 | 17 |
| 375 | 235.82 | temperature | 10 | 1440 | > th | dose dependent | rect. | 10 | 20 |
| 628 | 236.41 | temperature | 30 | 1440 | < th | dose dependent | rect. | 10 | 21 |
| 648 | 236.46 | temperature | 25 | 4320 | > th | dose independent | rect. | 20 | 6 |
| 684 | 236.53 | temperature | 20 | 1440 | > th | dose dependent | rect. | 5 | 2 |
| 719 | 236.59 | temperature | 10 | 1440 | > th | dose dependent | sin | 6 | NA |
| 726 | 236.59 | temperature | 30 | 270 | < th | dose dependent | rect. | 12 | 23 |
| 732 | 236.60 | temperature | 30 | 1440 | < th | dose dependent | rect. | 18 | 21 |
| 735 | 236.61 | temperature | 10 | 1440 | > th | dose dependent | no | NA | NA |
| 919 | 236.81 | temperature | 10 | 1440 | > th | dose dependent | rect. | 5 | 23 |
| 990 | 236.87 | temperature | 30 | 270 | < th | dose dependent | no | NA | NA |
| 995 | 236.88 | temperature | 20 | 4320 | > th | dose independent | rect. | 20 | 4 |