Os01g0562400
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Peptidylprolyl isomerase, FKBP-type domain containing protein.
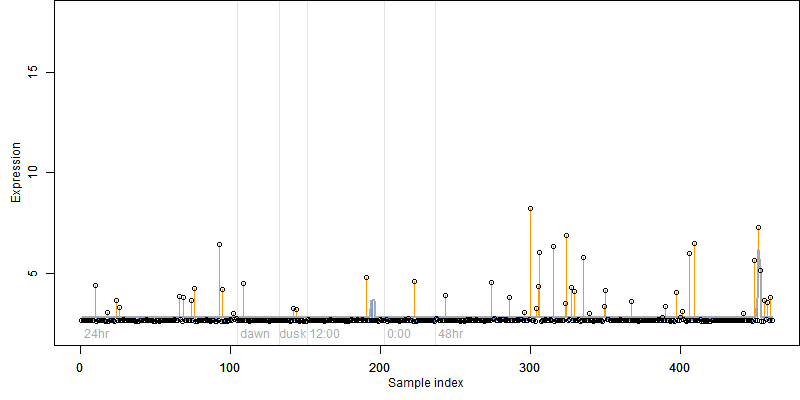

FiT-DB / Search/ Help/ Sample detail
|
Os01g0562400 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Peptidylprolyl isomerase, FKBP-type domain containing protein.
|
|
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__

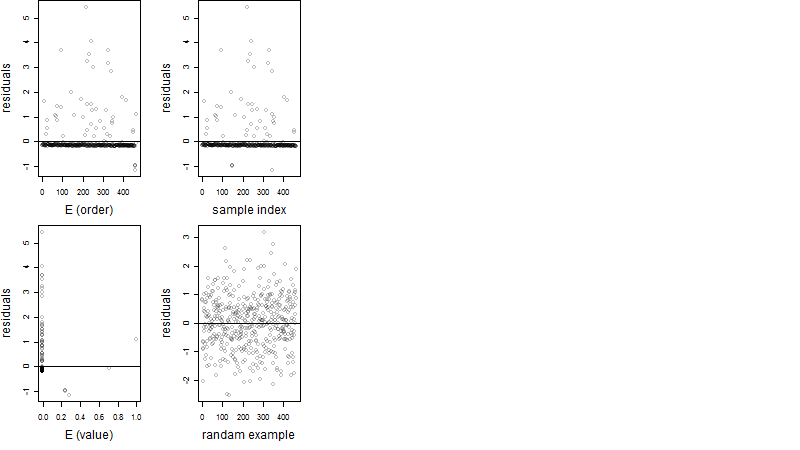
Dependence on each variable

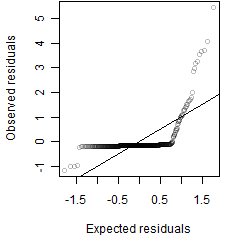
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = < th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 174.79 | 0.621 | 2.39 | 1.02 | 0.116 | -69.5 | 0.261 | 179 | -0.0326 | 6.94 | 45 | 231 | -- | -- |
| 198.27 | 0.661 | 2.81 | 0.1 | 0.0757 | -0.258 | 0.0159 | -- | -0.0406 | 10 | 45 | 231 | -- | -- |
| 174.95 | 0.616 | 2.39 | 1.03 | 0.117 | -69.5 | -- | 179 | -0.0333 | 7.37 | 45 | 232 | -- | -- |
| 180.36 | 0.625 | 2.86 | -0.169 | 0.0279 | 3.17 | -- | -- | -0.119 | 13.4 | 45 | 255 | -- | -- |
| 175.67 | 0.617 | 2.39 | 1.03 | -- | -69.5 | -- | 178 | -0.0315 | -- | 44.99 | 229 | -- | -- |
| 198.44 | 0.66 | 2.81 | 0.0926 | 0.0684 | -- | -0.132 | -- | -0.0405 | 8.25 | -- | -- | -- | -- |
| 198.46 | 0.66 | 2.81 | 0.0898 | 0.0711 | -- | -- | -- | -0.0398 | 9.69 | -- | -- | -- | -- |
| 178.93 | 0.623 | 2.81 | -0.0445 | -- | 3.41 | -- | -- | -0.0285 | -- | 44.72 | 242 | -- | -- |
| 178.98 | 0.623 | 2.81 | -- | 0.00242 | 3.39 | -- | -- | -0.0241 | 12.8 | 44 | 248 | -- | -- |
| 198.76 | 0.659 | 2.81 | 0.0892 | -- | -- | -- | -- | -0.0396 | -- | -- | -- | -- | -- |
| 198.67 | 0.659 | 2.81 | -- | 0.0709 | -- | -- | -- | -0.0498 | 9.6 | -- | -- | -- | -- |
| 178.98 | 0.623 | 2.81 | -- | -- | 3.38 | -- | -- | -0.0238 | -- | 44.52 | 243 | -- | -- |
| 198.97 | 0.658 | 2.82 | -- | -- | -- | -- | -- | -0.0495 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance
|
Histogram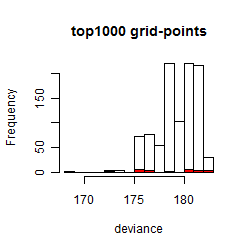 
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 168.98 | wind | 7 | 10 | > th | dose dependent | rect. | 12 | 1 |
| 3 | 172.84 | wind | 1 | 10 | < th | dose dependent | rect. | 14 | 4 |
| 11 | 175.02 | wind | 7 | 10 | > th | dose independent | rect. | 12 | 1 |
| 13 | 175.08 | wind | 1 | 10 | < th | dose independent | rect. | 14 | 4 |
| 21 | 175.39 | humidity | 50 | 270 | < th | dose independent | rect. | 14 | 1 |
| 22 | 175.53 | humidity | 50 | 270 | < th | dose dependent | rect. | 14 | 2 |
| 43 | 175.68 | humidity | 50 | 270 | < th | dose dependent | sin | 13 | NA |
| 84 | 176.02 | humidity | 50 | 270 | < th | dose independent | sin | 13 | NA |
| 87 | 176.21 | wind | 1 | 30 | < th | dose independent | rect. | 15 | 1 |
| 89 | 176.21 | wind | 1 | 30 | < th | dose dependent | rect. | 15 | 1 |
| 136 | 176.72 | humidity | 50 | 270 | > th | dose independent | sin | 13 | NA |
| 355 | 178.54 | humidity | 50 | 10 | < th | dose independent | rect. | 15 | 1 |
| 567 | 180.03 | humidity | 50 | 270 | < th | dose dependent | no | NA | NA |
| 695 | 180.38 | humidity | 90 | 90 | < th | dose dependent | rect. | 4 | 1 |
| 700 | 180.51 | wind | 7 | 1440 | > th | dose independent | rect. | 24 | 9 |
| 752 | 180.91 | wind | 7 | 1440 | > th | dose dependent | rect. | 24 | 9 |
| 753 | 180.95 | wind | 7 | 1440 | < th | dose independent | rect. | 24 | 9 |
| 776 | 181.24 | wind | 7 | 1440 | < th | dose independent | rect. | 22 | 11 |
| 832 | 181.93 | wind | 3 | 1440 | > th | dose dependent | rect. | 24 | 3 |
| 850 | 181.96 | humidity | 50 | 270 | < th | dose independent | no | NA | NA |
| 973 | 182.07 | wind | 5 | 1440 | < th | dose independent | rect. | 22 | 7 |
| 981 | 182.23 | wind | 5 | 1440 | > th | dose independent | rect. | 22 | 7 |
| 994 | 182.38 | wind | 5 | 1440 | < th | dose independent | rect. | 24 | 3 |