Os01g0223200
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
Description : Lipolytic enzyme, G-D-S-L family protein.

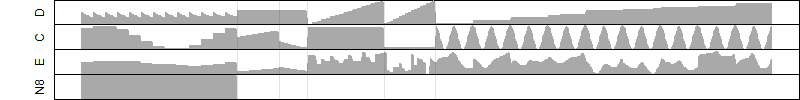
FiT-DB / Search/ Help/ Sample detail
|
Os01g0223200 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Lipolytic enzyme, G-D-S-L family protein.
|
|
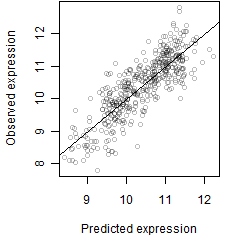
log2(Expression) ~ Norm(μ, σ2)
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|

 __
__
Dependence on each variable
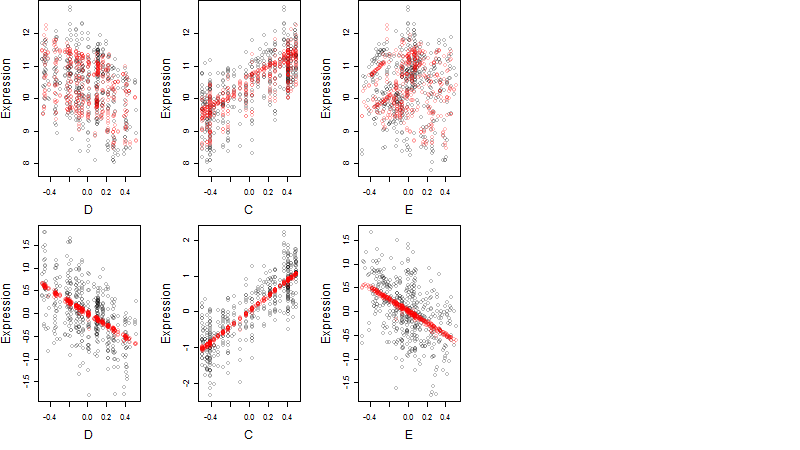
Residual plot

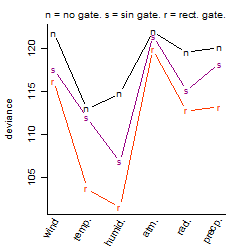
Process of the parameter reduction
(fixed parameters. wheather = humidity,
response mode = > th, dose dependency = dose dependent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 112.19 | 0.498 | 10.3 | -1.26 | 2.13 | -1.23 | 1.4 | -0.748 | 0.0245 | 9.83 | 53.09 | 1137 | -- | -- |
| 112.9 | 0.495 | 10.3 | -1.32 | 2.11 | -1.25 | 1.23 | -- | 0.0178 | 9.84 | 52 | 1149 | -- | -- |
| 118.72 | 0.507 | 10.3 | -1.28 | 2.12 | -1.23 | -- | 0.0216 | 0.0146 | 9.77 | 52.07 | 1138 | -- | -- |
| 118.72 | 0.507 | 10.3 | -1.28 | 2.11 | -1.22 | -- | -- | 0.017 | 9.76 | 52 | 1142 | -- | -- |
| 321.47 | 0.835 | 10.4 | -1.12 | -- | -1.26 | -- | -0.585 | -0.0601 | -- | 66 | 1496 | -- | -- |
| 143.34 | 0.561 | 10.3 | -1.26 | 1.87 | -- | 1.02 | -- | 0.062 | 9.43 | -- | -- | -- | -- |
| 147.12 | 0.568 | 10.3 | -1.22 | 1.87 | -- | -- | -- | 0.0625 | 9.37 | -- | -- | -- | -- |
| 321.76 | 0.835 | 10.4 | -1.22 | -- | -1.32 | -- | -- | -0.0605 | -- | 66 | 1495 | -- | -- |
| 160.16 | 0.589 | 10.3 | -- | 1.86 | -1.33 | -- | -- | 0.0629 | 9.58 | 71 | 1391 | -- | -- |
| 351.85 | 0.876 | 10.4 | -1.23 | -- | -- | -- | -- | 0.0695 | -- | -- | -- | -- | -- |
| 186.34 | 0.639 | 10.3 | -- | 1.87 | -- | -- | -- | 0.198 | 9.42 | -- | -- | -- | -- |
| 360.33 | 0.884 | 10.4 | -- | -- | -1.45 | -- | -- | 0.0323 | -- | 71 | 1500 | -- | -- |
| 391.75 | 0.924 | 10.4 | -- | -- | -- | -- | -- | 0.206 | -- | -- | -- | -- | -- |
Results of the grid search
Summarized heatmap of deviance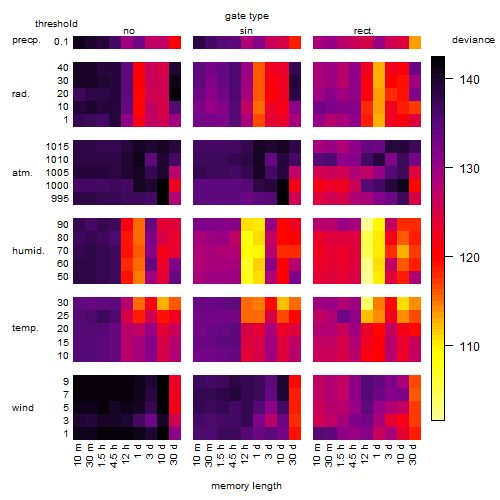
|
Histogram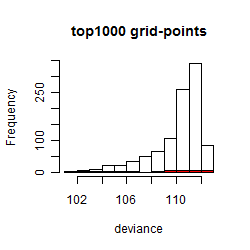 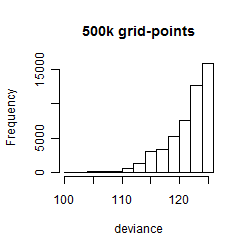
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 101.48 | humidity | 50 | 720 | > th | dose dependent | rect. | 16 | 11 |
| 3 | 101.99 | humidity | 90 | 720 | < th | dose dependent | rect. | 19 | 3 |
| 4 | 102.29 | humidity | 60 | 720 | > th | dose dependent | rect. | 18 | 1 |
| 15 | 103.68 | temperature | 30 | 720 | < th | dose independent | rect. | 17 | 2 |
| 16 | 103.87 | humidity | 60 | 720 | > th | dose dependent | rect. | 17 | 3 |
| 22 | 104.26 | humidity | 60 | 720 | > th | dose dependent | rect. | 16 | 5 |
| 27 | 104.39 | humidity | 70 | 720 | > th | dose independent | rect. | 16 | 11 |
| 31 | 104.78 | humidity | 70 | 720 | > th | dose independent | rect. | 16 | 4 |
| 42 | 105.11 | humidity | 80 | 720 | < th | dose independent | rect. | 19 | 2 |
| 48 | 105.37 | temperature | 30 | 720 | < th | dose independent | rect. | 17 | 10 |
| 85 | 106.78 | humidity | 60 | 720 | > th | dose dependent | sin | 16 | NA |
| 170 | 108.38 | humidity | 80 | 720 | > th | dose independent | sin | 17 | NA |
| 233 | 109.44 | humidity | 70 | 720 | > th | dose independent | sin | 13 | NA |
| 237 | 109.48 | humidity | 90 | 1440 | < th | dose dependent | rect. | 15 | 9 |
| 244 | 109.61 | humidity | 80 | 720 | > th | dose independent | rect. | 11 | 16 |
| 270 | 109.85 | humidity | 70 | 1440 | > th | dose independent | rect. | 14 | 17 |
| 316 | 110.01 | humidity | 90 | 1440 | < th | dose dependent | sin | 15 | NA |
| 324 | 110.13 | humidity | 70 | 720 | > th | dose independent | rect. | 16 | 17 |
| 459 | 110.77 | humidity | 80 | 720 | < th | dose independent | rect. | 11 | 11 |
| 469 | 110.79 | humidity | 70 | 1440 | < th | dose independent | sin | 15 | NA |
| 475 | 110.81 | humidity | 70 | 1440 | < th | dose independent | rect. | 14 | 6 |
| 488 | 110.81 | temperature | 30 | 14400 | > th | dose dependent | rect. | 17 | 2 |
| 635 | 111.25 | humidity | 50 | 720 | > th | dose dependent | rect. | 15 | 18 |
| 851 | 111.88 | temperature | 30 | 14400 | > th | dose independent | rect. | 17 | 2 |
| 899 | 111.96 | temperature | 30 | 14400 | > th | dose dependent | sin | 12 | NA |
| 903 | 111.97 | temperature | 30 | 14400 | < th | dose independent | rect. | 18 | 3 |
| 923 | 112.03 | temperature | 30 | 14400 | < th | dose independent | rect. | 18 | 10 |
| 937 | 112.06 | temperature | 30 | 14400 | < th | dose independent | rect. | 17 | 8 |
| 975 | 112.13 | temperature | 30 | 720 | < th | dose independent | rect. | 16 | 17 |
| 989 | 112.19 | temperature | 25 | 720 | > th | dose dependent | rect. | 19 | 3 |