Os01g0114100
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II
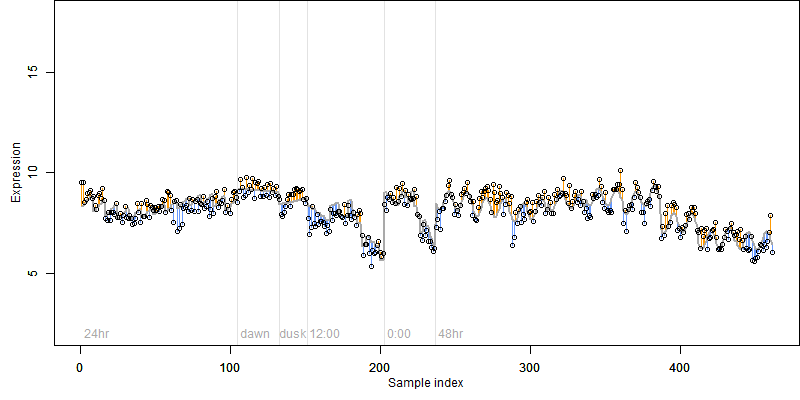
Description : Similar to Protein kinase RLK17.
FiT-DB / Search/ Help/ Sample detail
|
Os01g0114100 |
blastp(Os)/ blastp(At)/ coex/// RAP/ RiceXPro/ SALAD/ ATTED-II |
|
Description : Similar to Protein kinase RLK17.
|
|
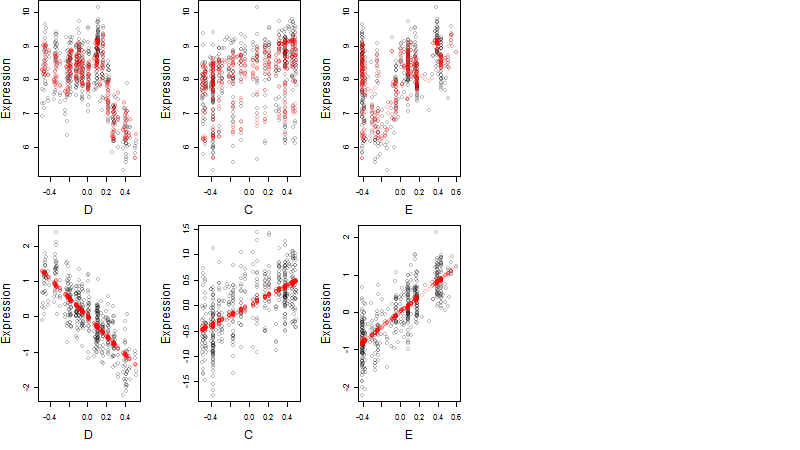
log2(Expression) ~ Norm(μ, σ2)
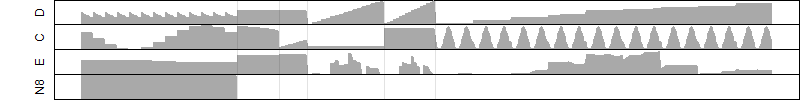
μ = α + β1D + β2C + β3E + β4D*C + β5D*E + γ1N8
|
_____ |
|
 __
__
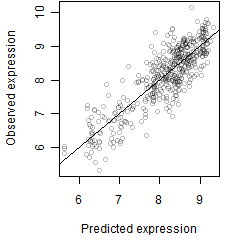
Dependence on each variable
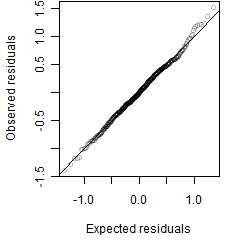
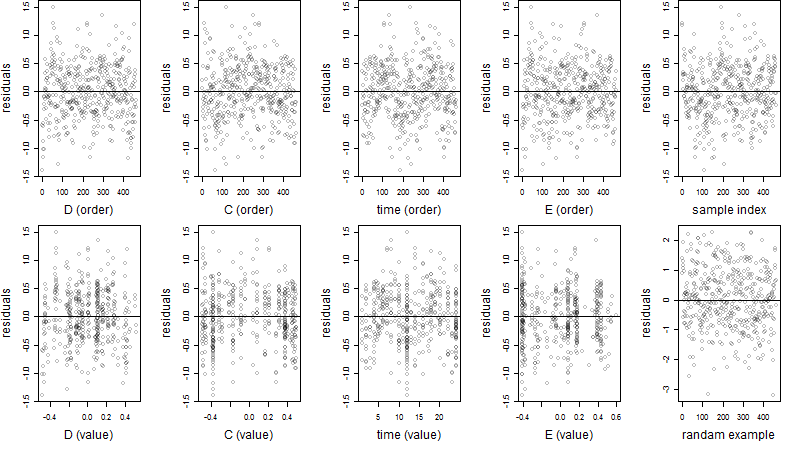
Residual plot
Process of the parameter reduction
(fixed parameters. wheather = temperature,
response mode = > th, dose dependency = dose independent, type of G = no)
| deviance | σ | α | β1 | β2 | β3 | β4 | β5 | γ1 | peak time of C | threshold | memory length | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 102.29 | 0.475 | 8.22 | -2.79 | 0.963 | 2.07 | -0.67 | -0.299 | -0.436 | 2.64 | 27.3 | 5910 | -- | -- |
| 102.37 | 0.471 | 8.21 | -2.72 | 0.954 | 2.04 | -0.662 | -- | -0.419 | 2.62 | 27.4 | 5920 | -- | -- |
| 103.87 | 0.475 | 8.22 | -2.75 | 0.965 | 2.06 | -- | -0.221 | -0.429 | 2.67 | 27.36 | 5920 | -- | -- |
| 103.9 | 0.475 | 8.21 | -2.68 | 0.964 | 2.04 | -- | -- | -0.406 | 2.68 | 27.34 | 5920 | -- | -- |
| 148.6 | 0.568 | 8.22 | -2.66 | -- | 2.08 | -- | 0.326 | -0.502 | -- | 25.8 | 6922 | -- | -- |
| 251.02 | 0.743 | 8.11 | -1.85 | 0.998 | -- | -0.403 | -- | 0.0346 | 2.57 | -- | -- | -- | -- |
| 251.6 | 0.743 | 8.11 | -1.84 | 0.997 | -- | -- | -- | 0.0369 | 2.59 | -- | -- | -- | -- |
| 145.85 | 0.562 | 8.19 | -2.66 | -- | 2.18 | -- | -- | -0.328 | -- | 27.4 | 5492 | -- | -- |
| 258.33 | 0.749 | 8.09 | -- | 1.01 | 1.36 | -- | -- | 0.204 | 2.17 | 29.25 | 6054 | -- | -- |
| 309.86 | 0.823 | 8.1 | -1.8 | -- | -- | -- | -- | 0.062 | -- | -- | -- | -- | -- |
| 340.75 | 0.863 | 8.06 | -- | 0.962 | -- | -- | -- | 0.242 | 2.56 | -- | -- | -- | -- |
| 295.43 | 0.801 | 8.06 | -- | -- | 1.51 | -- | -- | 0.266 | -- | 29.3 | 5479 | -- | -- |
| 395.01 | 0.928 | 8.06 | -- | -- | -- | -- | -- | 0.261 | -- | -- | -- | -- | -- |
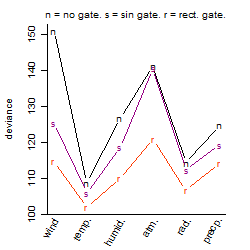
Results of the grid search
Summarized heatmap of deviance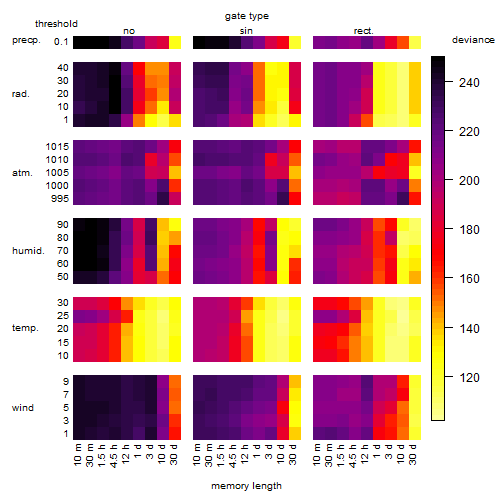
|
Histogram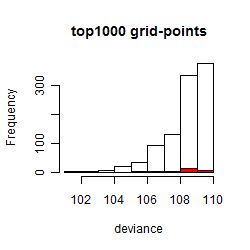 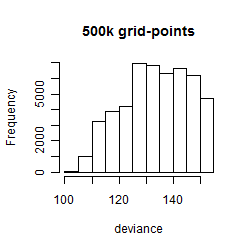
|
Local optima within top1000 grid-points
| rank | deviance | wheather | threshold | memory length | response mode | dose dependency | type of G | peak or start time of G | open length of G |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 101.94 | temperature | 25 | 14400 | > th | dose dependent | rect. | 17 | 1 |
| 50 | 105.47 | temperature | 30 | 14400 | < th | dose independent | rect. | 15 | 1 |
| 57 | 105.81 | temperature | 30 | 14400 | > th | dose independent | rect. | 15 | 1 |
| 58 | 105.86 | temperature | 25 | 14400 | > th | dose dependent | sin | 16 | NA |
| 103 | 106.65 | radiation | 1 | 14400 | > th | dose independent | rect. | 18 | 1 |
| 126 | 106.77 | temperature | 30 | 14400 | < th | dose independent | rect. | 14 | 14 |
| 135 | 106.79 | temperature | 30 | 14400 | < th | dose independent | rect. | 14 | 11 |
| 182 | 107.19 | radiation | 1 | 14400 | < th | dose independent | rect. | 18 | 10 |
| 210 | 107.43 | radiation | 1 | 14400 | < th | dose independent | rect. | 18 | 1 |
| 213 | 107.45 | radiation | 1 | 14400 | < th | dose independent | rect. | 18 | 3 |
| 387 | 108.44 | temperature | 25 | 14400 | < th | dose independent | sin | 15 | NA |
| 392 | 108.47 | temperature | 10 | 4320 | > th | dose dependent | rect. | 6 | 3 |
| 397 | 108.51 | radiation | 10 | 14400 | < th | dose dependent | rect. | 17 | 12 |
| 399 | 108.53 | temperature | 25 | 14400 | > th | dose independent | sin | 15 | NA |
| 412 | 108.59 | temperature | 25 | 14400 | > th | dose dependent | rect. | 3 | 21 |
| 436 | 108.64 | temperature | 25 | 14400 | > th | dose dependent | rect. | 21 | 23 |
| 458 | 108.68 | temperature | 30 | 4320 | < th | dose dependent | rect. | 6 | 3 |
| 467 | 108.69 | temperature | 25 | 14400 | > th | dose dependent | no | NA | NA |
| 529 | 108.77 | radiation | 40 | 14400 | < th | dose dependent | rect. | 17 | 12 |
| 534 | 108.78 | radiation | 10 | 14400 | < th | dose dependent | rect. | 17 | 7 |
| 544 | 108.80 | radiation | 10 | 14400 | < th | dose dependent | rect. | 17 | 2 |
| 550 | 108.81 | temperature | 30 | 14400 | < th | dose dependent | rect. | 18 | 1 |
| 816 | 109.51 | temperature | 10 | 4320 | > th | dose dependent | rect. | 7 | 1 |
| 899 | 109.71 | temperature | 30 | 4320 | < th | dose dependent | rect. | 7 | 1 |
| 954 | 109.85 | radiation | 1 | 14400 | > th | dose dependent | rect. | 17 | 12 |
| 957 | 109.86 | humidity | 90 | 43200 | < th | dose independent | rect. | 17 | 2 |
| 992 | 109.94 | radiation | 1 | 14400 | > th | dose dependent | rect. | 17 | 2 |